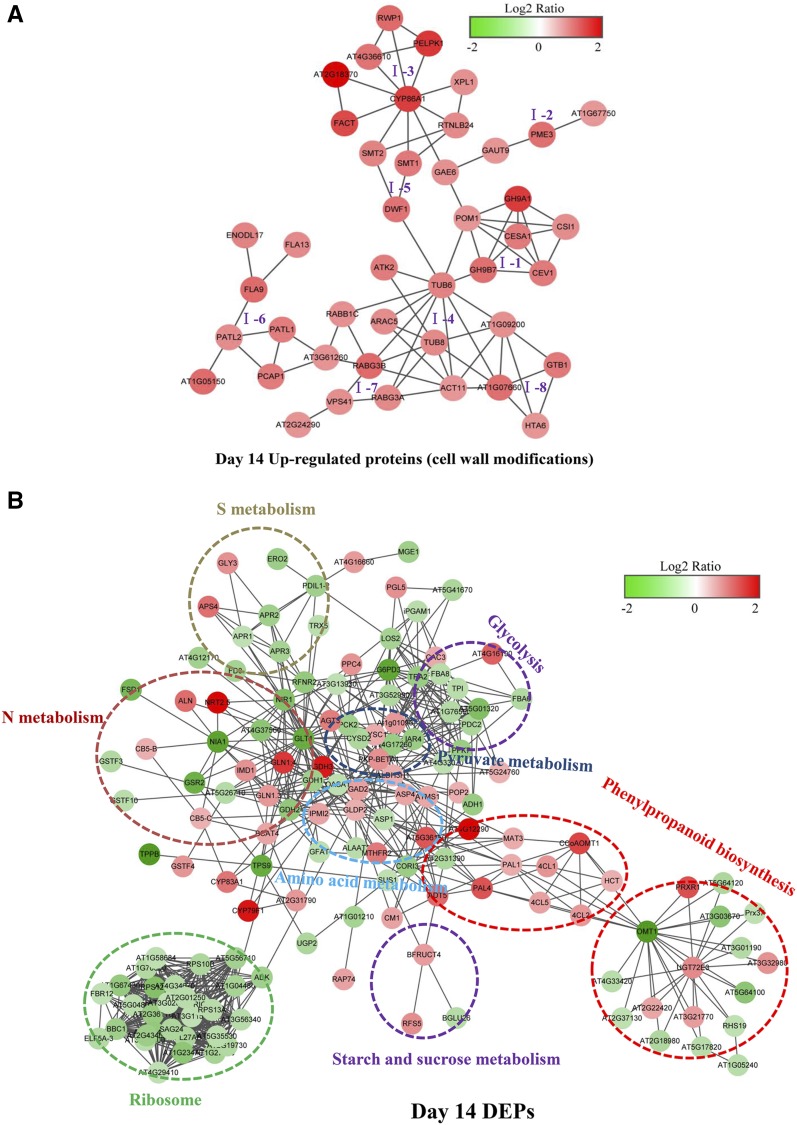
Figure 6.
Predicted protein-protein interaction networks of DEPs in day-14 N-deficient rapeseed roots. A, Predicted protein-protein interaction networks of DEPs (up-regulated proteins) involved in cell wall metabolism in day-14 N-deficient rapeseed roots. Eight subclusters are grouped according to the pathways connected with these proteins: I-1, cellulose synthesis; I-2, pectin metabolism; I-3, cutin, suberin, and wax biosynthesis; I-4, cytoskeleton organization; I-5, sterol composition; I-6, membrane trafficking; I-7, vacuolar trafficking; and I-8, signal transduction and transcription. B, Predicted protein-protein interaction networks of all DEPs, except for those involved in cell wall metabolism, in day-14 N-deficient roots. DEPs within dotted circles share common metabolic pathways. The intensity of color in the circles indicates the degree of up-regulation (red) or down-regulation (green) of the DEPs. This interaction network was constructed using the STRING protein-protein network analysis program (http://string-db.org/) with the Arabidopsis database and confidence scores greater than 0.7. Network files were downloaded from STRING, and network images were made in Cytoscape 3.3.0. The full names for all the DEPs shown in the protein-protein interaction networks are listed in Supplemental Table S3.