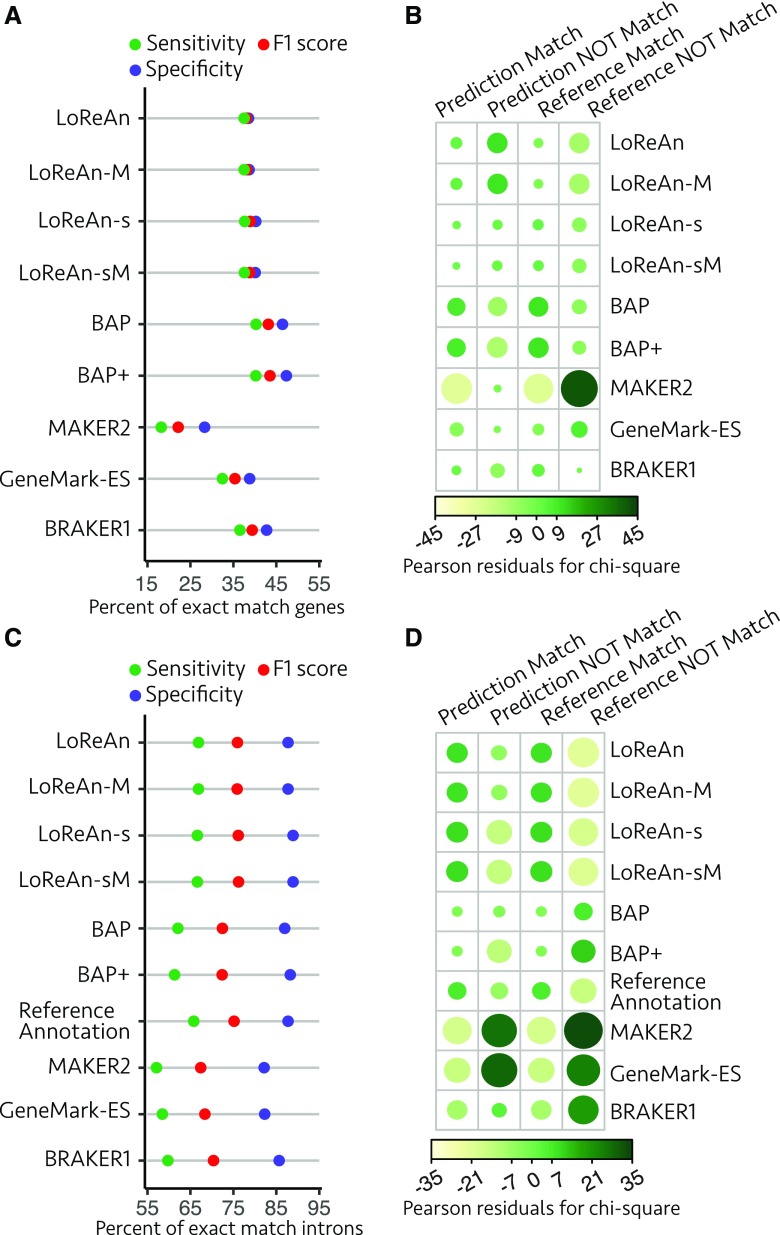
Figure 6.
Assessment of gene and intron predictions from P. crispa. A, Annotation quality metrics are shown for exact match genes as detailed for Figure 2A. LoReAn, LoReAn in nonstranded mode using a nonmasked genome; LoReAn-M, LoReAn in nonstranded mode using a masked genome; LoReAn-s, LoReAn in stranded mode using a nonmasked genome; LoReAn-sM, LoReAn in stranded mode using a masked genome; BAP, broad annotation pipeline; BAP+, broad annotation pipeline plus additional modifications described in text. B, The proportion of exact match to nonmatching gene predictions (specificity) and exact match to missing gene predictions (sensitivity) compared using a χ2 test of independence as described in Figure 2B. C and D are for exact match intron analysis, represented as in A and B, respectively.