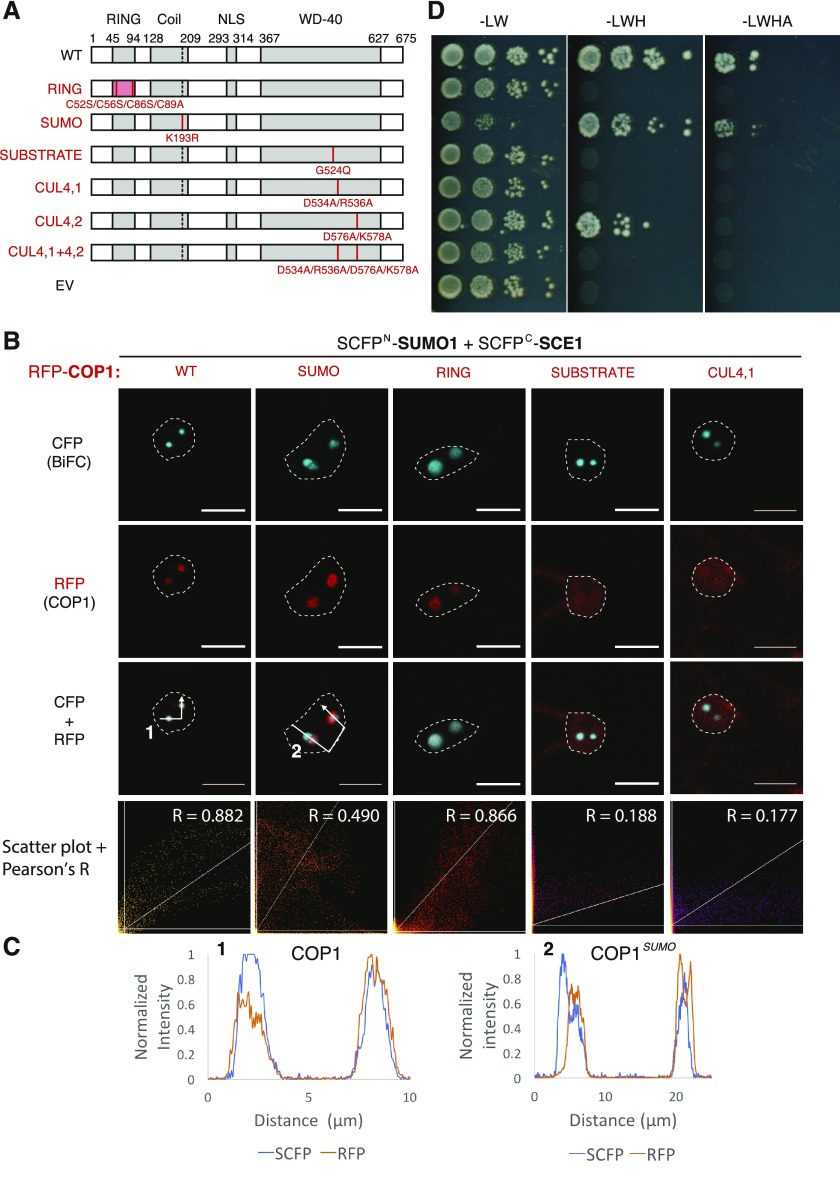
Figure 3.
The COP1-SIZ1 interaction in NBs depends on both the substrate binding pocket and the SUMO acceptor site in COP1. A, Schematic representation of COP1 variants and the protein-protein interaction domain disrupted (left side, red text) with their mutations shown: RING, RING Zn2+-finger binding domain mutant; SUMO, loss of SUMO acceptor site; SUBSTRATE, mutation in the substrate binding groove; CUL4, mutations in the CUL4-binding “WDRX” motifs. WT, Wild type. B, Nuclear localization pattern of the SUMO1∙SCE1 BiFC pair in cells overexpressing functional mutants of RFP-COP1. Micrographs depict from top to bottom the BiFC CFP signal, RFP-COP1, and their combined signals. Bottom row depicts a scatter plot of the RFP/CFP signal intensities per pixel and their Pearson’s correlation coefficient (R). Conditions were identical to those in Figure 1. Scale bars, 10 µm. C, Normalized intensity profiles depict the fluorescence signal intensities for SCFP: (BiFC pair) and RFP-COP1: coexpression of (1) wild-type COP1 or (2) COP1SUMO; the profiles follow the white arrows depicted in B. Note the profiles of RFP-COP1SUMO (A) and SUMO∙SCE1 CFP signals poorly overlap in (2). D, Mapping of the SIZ1 interaction site in COP1 using Y2H analysis of the COP1 variants depicted in B. The COP1 variants were fused to the GAL4 BD, whereas SIZ1 was fused to the GAL4 AD-fusion. Yeast growth was scored 3 d after incubation on selective medium at 30°C.