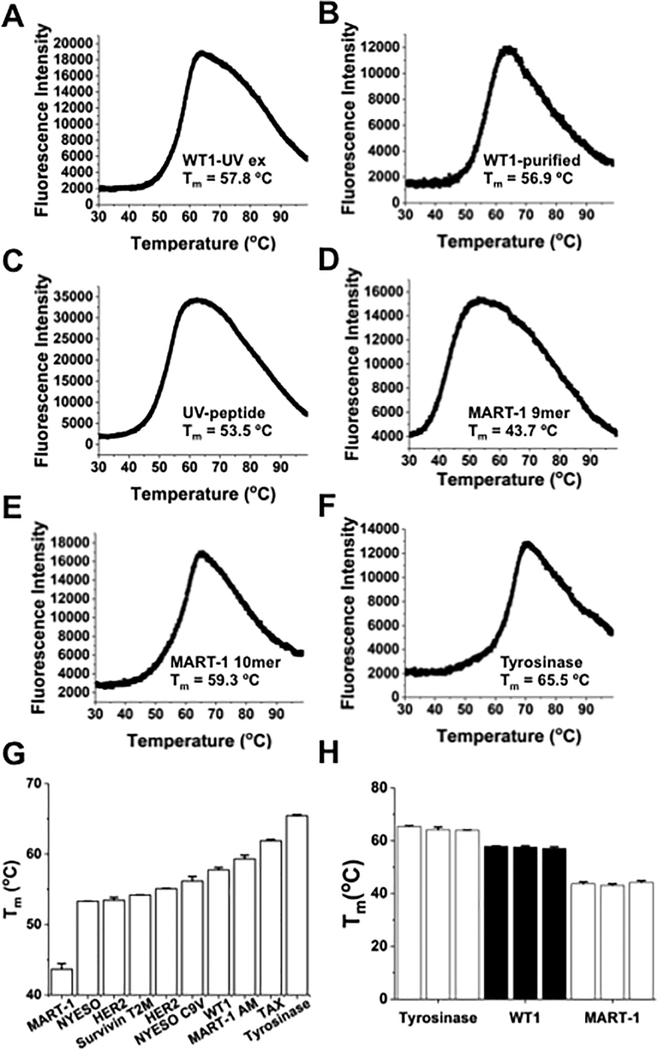
Figure 1. Thermal denaturation curves generated from differential scanning fluorimetry (DSF) of self-peptide/HLA-A2 complexes.
(A) DSF denaturation profile of WT-1 (RMFPNAPYL)/HLA-A2 complex prepared through UV-mediated ligand exchange at 4 uM. For all measurements, the temperature ramp rate was 1°C/minute from 25°C to 99°C. The curve was created using OriginPro 2017 software to plot all 820 time points generated from QuantStudio™ Real-Time PCR. (B) Thermal denaturation curve of refolded WT-1/HLA-A2 monomer at 4 uM, used for comparison with UV-generated complex. (C) DSF profile of refolded UV-peptide/HLA-A2. The UV-peptide (KILGFVFJV) contains 3-amino-3-(2-nitro)phenyl-propionic acid, denoted with a J in the peptide sequence. (D-F) DSF profiles for UV-exchanged self-peptide/HLA-A2 complexes with MART-1 9-mer (AAGIGILTV), MART-1 anchor-modified 10-mer (ELAGIGILTV), and Tyrosinase (YMDGTMSQV). For (A-F), curves are representative of 3 independent experiments, with three replicates per experiment. (G) Comparison of Tm values from several well-characterized self-peptides, including NYESO-1 (SLLMWITNC) and HER2 (KIFGSLAFL). (H) Comparison of Tm values from three separate DSF experiments with UV-exchanged self-peptide/HLA-A2 complexes. UV-exchanged complexes with Tyrosinase (YMDGTMSQV), WT-1 (RMFPNAPYL), and MART-1 (AAGIGILTV) were analyzed in three separate experiments of three replicates each. Different UV-exchanged preparations were used for each trial. Error bars represent the standard deviation between replicates of individual DSF runs.