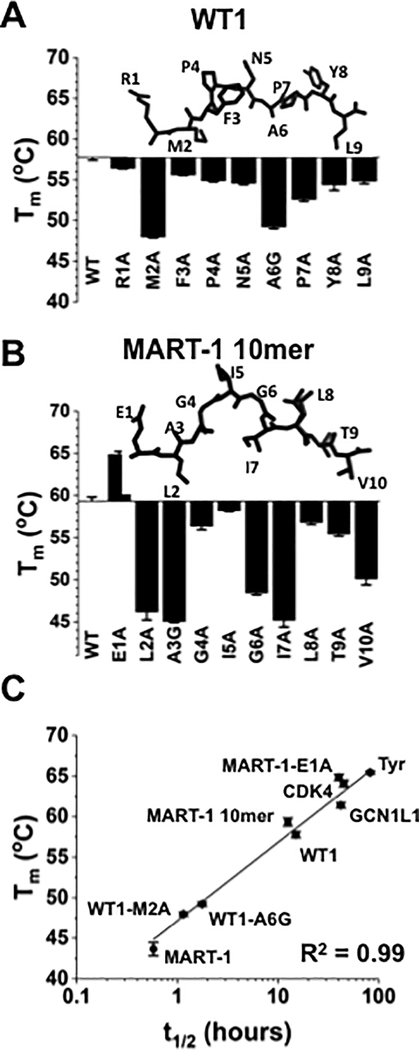
Figure 4. DSF analysis of self-peptides and single peptide variants.
(A) Comparison of Tm values for self-peptide WT-1 (RMFPNAPYL) and its alanine variants. In positions where an alanine was present in the wild-type sequence, it was replaced with a glycine (WT1-A6G). The structure of the WT-1 peptide from the WT-1/HLA-A2 structure is shown (PDB: 3HPJ). (B) Tm values for MART-1 anchor-modified 10-mer (ELAGIGILTV) and its alanine variants. In positions where an alanine was present in the wild-type sequence, it was replaced with a glycine (MART1-A3G). The structure of the MART-1 10mer peptide from the MART-1/HLA-A2 structure is shown (PDB: 1JF1). (C) Correlation (R2=0.99) between half-lives measured experimentally using the anti-β2m probe and Tm values obtained from DSF. Error bars represent the average and standard deviation of two independent experiments, with three replicates per experiment.