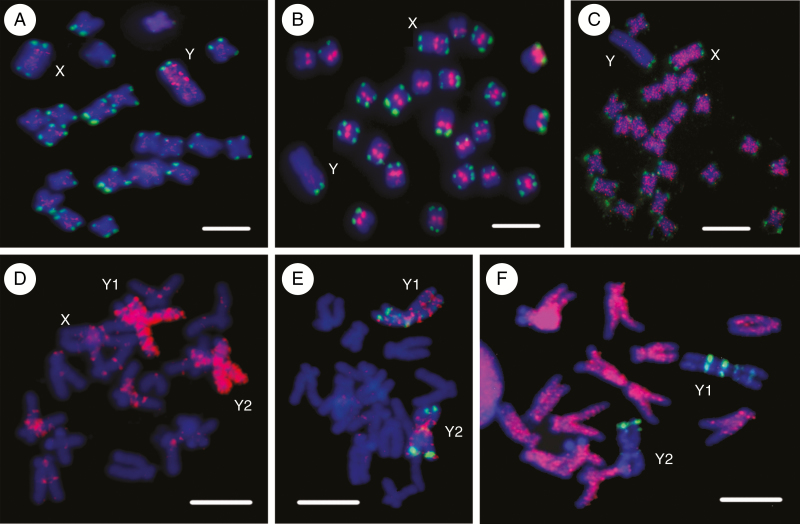
Fig. 4.
Chromosomal localization of repetitive DNA in Silene latifolia (A–C) and Rumex acetosa (D–F) determined by fluorescence in situ hybridization. (A) Microsatellite (CA)15 is accumulated on the Y chromosome (red signal), (B) satellite STAR (red signal) is present in the centromeres of all autosomes and the X chromosome, and satellite X-43.1 (green signal) is gathered in both sub-telomeres of all chromosomes with the exception of the Y chromosome possessing only one sub-telomeric signal, (C) The Ogre LTR retrotransposon (red signal) is ubiquitous on all chromosomes except the Y chromosome. (D) A mixture of all mono-, di- and trinucleotide microsatellites (red signal) shows strong accumulation along both Y chromosomes. (E) The (TA)15 microsatellite (red signal) gives a signal at several discrete loci on both Y chromosomes, and the RAYSI satellite (green signal) is present at the distal regions of both Y chromosomes. (F) The Maximus/SIRE LTR retrotransposon (red signal) covers all autosomes and the X chromosome except telomeres/sub-telomeres, but is absent from both Y chromosomes; the RAYSI satellite is localized at several discrete loci on both Y chromosomes (green signal). Scale bars indicate 10 μm.