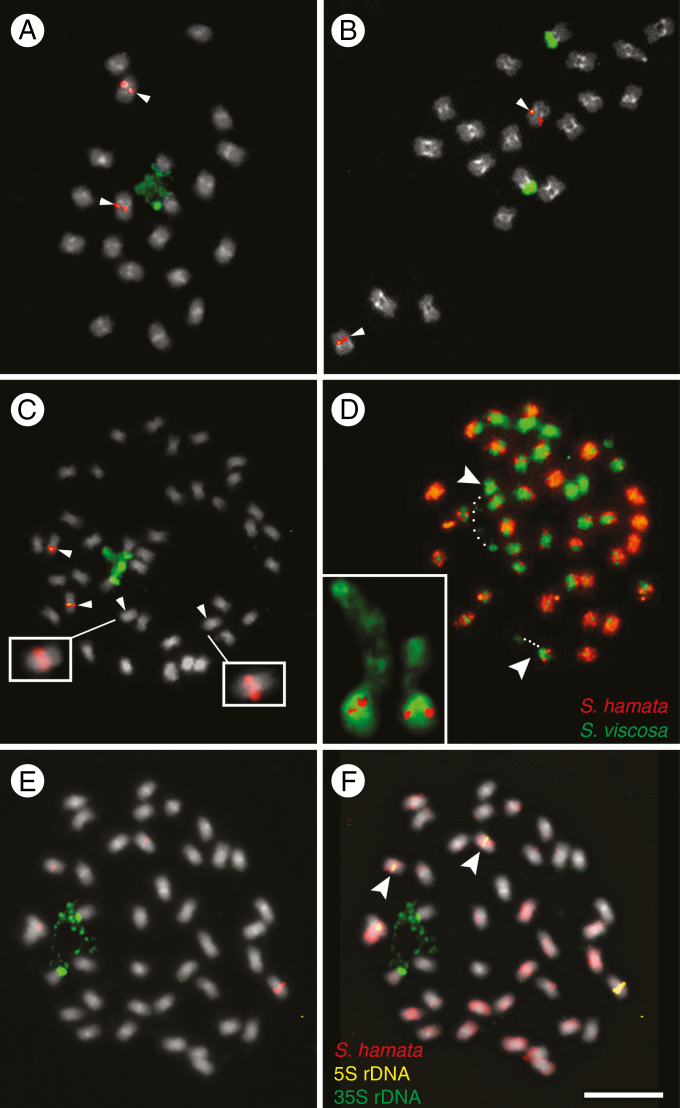
Fig. 1.
FISH and GISH in the Stylosanthes scabra complex. (A–C, E) FISH with 5S (red) and 35S (green) rDNA probes in S. viscosa (A), S. hamata (B) and S. scabra (C, E). Arrowheads in (C) show chromosomes harbouring the 5S rDNA. Insets in (C) show overexposed S. scabra chromosomes with weak 5S rDNA signals. (D) GISH in S. scabra using the genomic probes of S. hamata (orange) and S. viscosa (green). (F) GISH in S. scabra using the gDNA of S. hamata as probe (red) and the gDNA of S. viscosa as blocking DNA (grey). (F) The same cell as in (E) after sequential rDNA-FISH and GISH. Arrowheads and the inset in (D) show the NOR-harbouring chromosomes labelled with S. viscosa probe in green. Note the position of assumed decondensed 35S rDNA (dotted lines) connecting a satellite to the rest of the chromosome (dotted lines). Note also that in (F) the 35S rDNA-harbouring chromosomes are not labelled with the S. hamata probe, while the chromosome pair with the weak 5S rDNA signal is labelled in red (arrowheads).