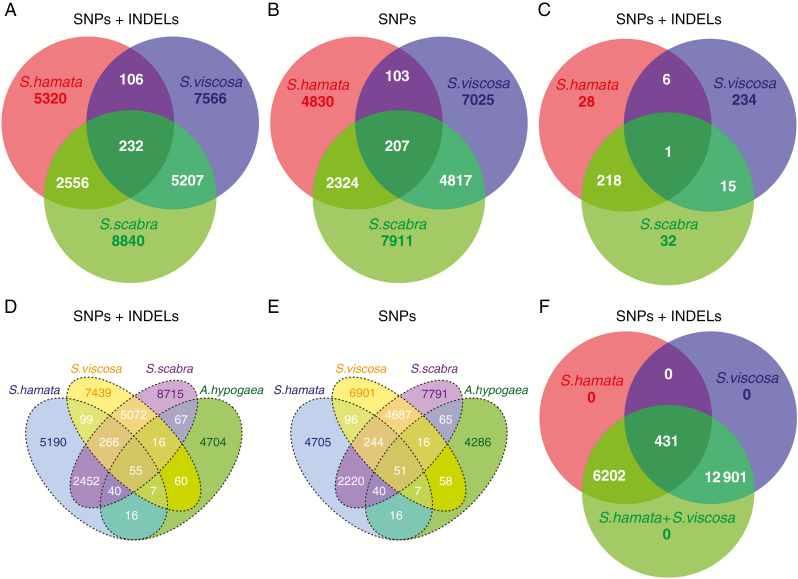
Fig. 6.
Venn diagram showing the amount of genome-wide reference-free shared and unique SNVs detected in each Stylosanthes genome. (A) Genome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and S. scabra. (B) SNP only analysis with S. hamata, S. viscosa and S. scabra. (C) Plastome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and S. scabra. (D) Analysis of SNPs + INDELs with S. hamata, S. viscosa, S. scabra and A. hypogaea. (E) SNP only analysis with S. hamata, S. viscosa, S. scabra and A. hypogaea. (F) Genome-wide analysis of SNPs + INDELs with S. hamata, S. viscosa and a virtual hybrid (S. hamata + S.viscosa) as control.