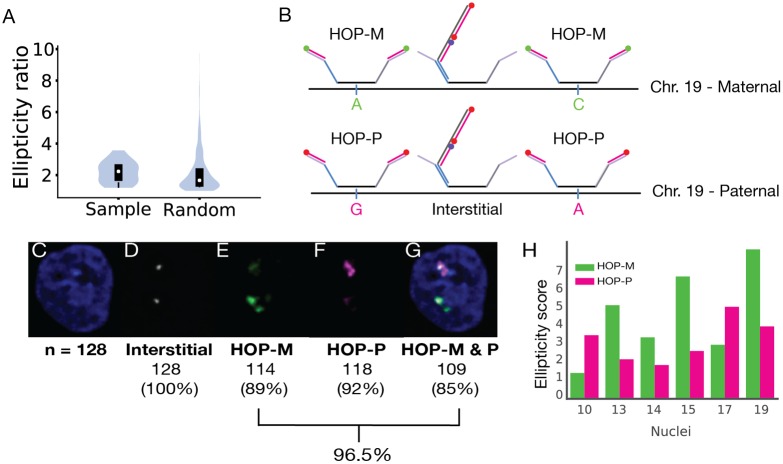
Fig 5. Homolog-specific Oligopaints for PGP1f.
(A) Violin plots of the ratios of 19 pairs of ellipticity scores, each pair representing the two homologs of one of the 19 imaged nuclei (Sample) or of 1,000 pairs of ellipticity scores, each pair representing two chromosomes chosen at random from the 38 representing the 19 imaged nuclei (Random). Boundaries of the black box-plot represent 1st and 3rd quartiles, white dot represents the median, and whiskers extend to 1.5 times the interquartile range (Mann-Whitney rank test, *: p < 10−2). (B) The HOP-M and HOP-P probes for chromosome 19 each encompass the entire chromosome and contain 11,259 oligos that cover ≥ 1 SNV per oligo. Thus, they are in contrast to Oligopaint oligos used to image CS1-9, these latter probes being “interstitial” in nature, as they avoid SNVs. HOP-M and HOP-P probes are visualized with secondary oligos labeled with different dyes, such that the two probes can be distinguished. (C-G) Images of a nucleus visualized with DAPI (C, blue), an interstitial probe targeting just CS3 (D, grey), HOP-M (E, green, Atto488N), HOP-P (F, magenta, Atto565N), and all probes (G), the latter demonstrating co-localization of all signals (n = 128); percentages show efficiency of each probe configuration, with a combined efficiency of 96.5%. (H) Ellipticity for the maternal (green) and paternal (magenta) homologs of the 6 nuclei for which HOPs had been applied.