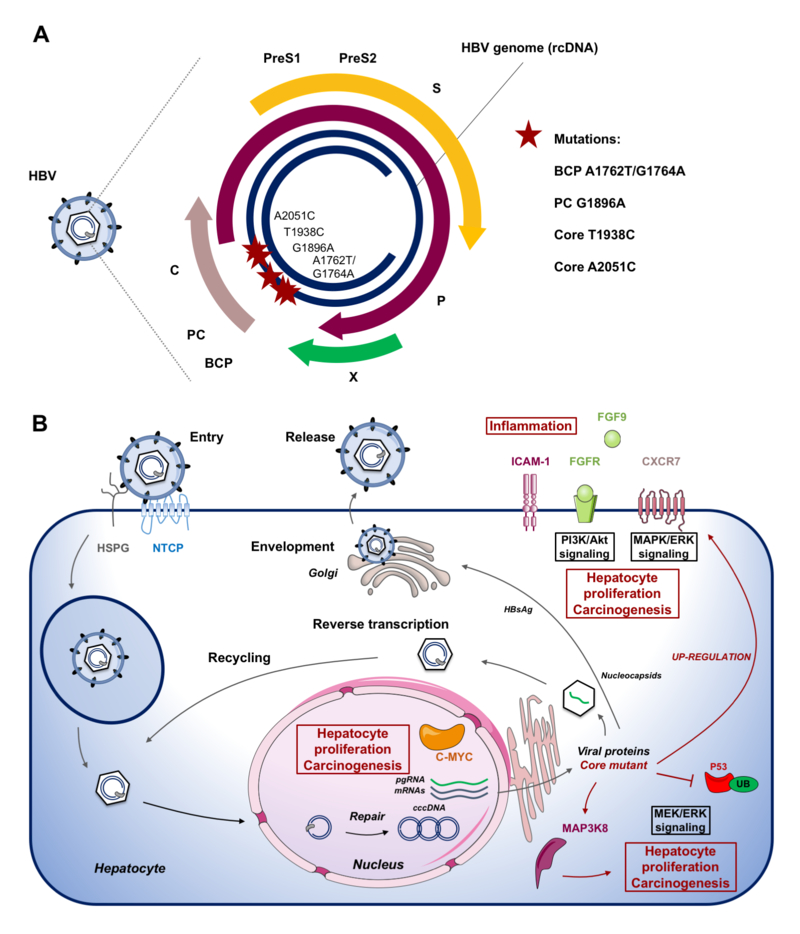
Figure 1: HBV core mutations associated with fibrosis and hepatocarcinogenesis.
A. Schematic representation of the HBV genome with clinical mutations. An accumulation of Core T1938C and A2051C mutations together with BCP (A1762T/G1764A) and PC (G1896A) mutations were observed in Alaskan native HCC patients (9). The mutations, leading to an increased viral replication, are indicated by stars on the genome. B. Effects of HBV core mutants on the HBV life cycle and host cell pathways in liver chimeric mice as observed by Hayashi et al. (9). Following viral entry mediated by HSPG and NTCP, the HBV genome is released into the nucleus where rcDNA is converted into cccDNA. cccDNA serves as template for viral transcripts including pgRNA, which is encapsidated and reverse transcribed into de novo HBV genomic DNA before assembly and release of the viral particle. Hayashi et al. demonstrated that core mutations enhance HBV replication in genotype F1b. In human liver chimeric mice infection with mutant virus resulted in up-regulation/activation of pathways potentially driving fibrosis and carcinogenesis. Abbreviations: BCP, basal core promotor; c-MYC, c-myc avian myelocytomatosis viral oncogene homolog; cccDNA, covalently closed circular DNA; CXCR7, C-X-C chemokine receptor type 7; FGF9, fibroblast growth factor 9; FGFR, fibroblast growth factor receptor; ICAM-1, intercellular adhesion molecule 1; MAP3K8, mitogen-activated protein; NTCP, NA+-taurocholate costransporting polypeptide; P, polymerase; HSPG, heparan sulfate proteoglycan; PC, core promotor; pgRNA, pregenome RNA; PreS, PreS region; S, surface protein; X, X protein.