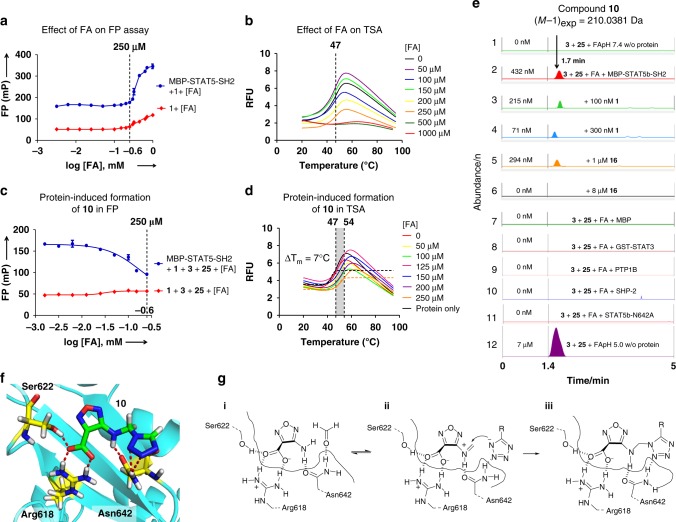
Fig. 3.
Assembly of STAT5 inhibitor 10 through protein-induced Mannich ligations. a FA was tolerated at up to 250 µm in the FP assay of MBP-STAT5b-SH2 (n = 3). b FA did not affect the STAT5b protein stability in the TSA at ≤ 250 µm (data shown are one representative of n = 3). c Protein-induced formation of 10 in the FP assay with increasing FA concentrations (n = 3). d Protein-induced formation of 10 from fragments 3 and 1H-tetrazole 25 with increasing FA concentrations in the TSA (ΔTm = 7 °C) (data shown are one representative of n = 3). e Formation of 10 detected in the HPLC-QTOF-MS. 1: No formation of 10 from FA, fragments 3 and 25 (all 250 µm) at pH 7.4 in MOPS buffer without protein. 2: Protein-induced formation of compound 10 with protein MBP-STAT5b-SH2 (250 nm) at pH 7.4. 3–6: Inhibition of protein-induced formation of 10 by peptide 1 (3,4) or inhibitor 16 (5,6). 7,8: No formation of compound 10 in the presence of Maltose Binding Protein (1 µm) or GST-STAT3 (250 nm). 9–11: Compound 10 was not formed in the presence of phosphatases, SHP2 (250 nm) and PTP1B (250 nm) nor in the presence of mutant STAT5b-N642A at pH 7.4 in MOPS buffer. 12: Formation of compound 10 at pH 5.0 without protein. Data shown are one representative example of n = 3. f 3D-binding model of the STAT5b:10 complex. Hydrogen bonds are illustrated as red dashed lines. g, Mechanism of the protein-induced formation of 10 from 3 with FA and 25 (R = H). (i) Binding of 3 via Arg618, Ser622, and Asn642, activation of FA with Asn642. (ii) Activation of the forminium cation of 3, coordination of the incoming tetrazolium anion of 25 by Asn642 leads to formation of Mannich ligation product 10 (iii). Error bars denote mean ± S.D