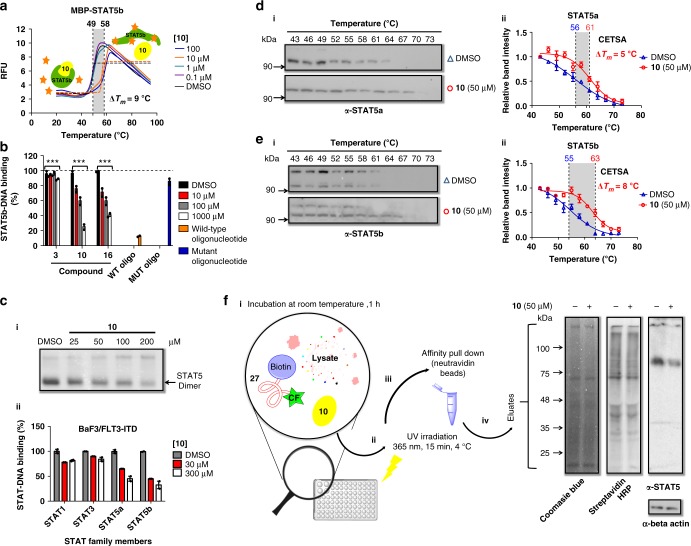
Fig. 4.
Inhibitor 10 blocks STAT5 dimerization, DNA-binding, and is active in CETSA and photocrosslinking assays in cellular lysates. a 10 stabilizes MBP-STAT5b-SH2 shifting the melting temperature (ΔTm) by 9 °C in the TSA (data shown are one representative example of n = 3). b 10 and 16 inhibit formation of trimeric (STAT5b)2-DNA complexes in an ELISA (n = 3). c (i) 10 inhibits binding of STAT5 dimers, isolated from nuclear extracts of BaF3/FLT3-ITD cells, to its target DNA in the electro mobility shift assay (EMSA) and (ii) shows selectivity for disrupting STAT5a,b-DNA complexes in the TransAM® STAT family ELISA (n = 3). d–e 10 binds to STAT5a and STAT5b in complex cellular lysates as demonstrated by cellular TSA (CETSA), resulting in shifted melting curves at 50 µm compared with vehicle (DMSO). Relative STAT5a and STAT5b band intensities were plotted against corresponding incubation temperatures and fitted to Boltzmann sigmoidal curve. The blue triangle represents DMSO and red circle represents compound 10 (n = 3). f Binding of 10 to STAT5 in BaF3/FLT3-ITD cell lysate was determined by photo-crosslinking (n = 3). (i) The dual-labeled (carboxyfluorescein and biotin) peptide probe 27 binds to STAT5 with submicromolar affinity and photocrosslinked (ii) with target proteins by activating the 4-phosphoncarbonyl residue that acts as a photoactive phosphotyrosine-mimetic. (iii) Cross-linked proteins can be isolated by biotin pull-down using Neutravidine beads. (iv) Displacement of 27 (100 µm) by compound 10 (50 µm) in BaF3/FLT3-ITD cell lysates (1 mg ml−1) resulted in significantly reduced photo-crosslinking of 27 and STAT5 as demonstrated in the western blotting using STAT5 antibodies (right lane), whereas other biotinylated proteins were not reduced (middle lane). For an uncropped image of c, d, e, f see Supplementary Figure 10. Error bars denote mean ± S.D. and p values are considered as follows: *p value < 0.05; **p value < 0.01; and ***p value < 0.001. Statistical analyses were performed using one-way ANOVA or the two-tailed Student’s t tests where appropriate