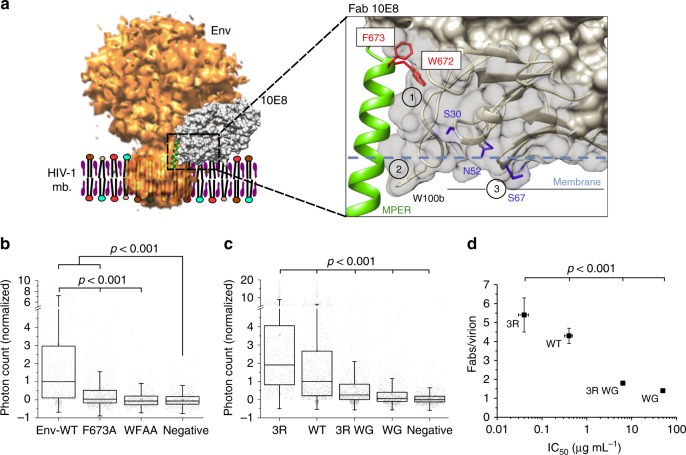
Fig. 5.
Dependence of 10E8 binding to native MPER on the functional paratope elements. a Structural model to designate: (1) specificity pocket, (2) HCDR3, and (3) MAPA regions on 10E8 (EMD entry code 330815 and PDB 5GHW10). Side chains of residues Trp672 and Phe673 critical for epitope peptide binding and neutralization are displayed in red onto the bound MPER helix (green ribbon). Fab residues from the HCDR3/MAPA region modulating membrane interaction and neutralization activity, Trp100bHC, and residues mutated to Arg (in blue: Ser30LC, Asn52LC, Ser67LC) are displayed in stick representation. b STED intensity signals of the antibodies (after background subtraction and normalization) in HIV-1 virions pseudotyped with JR-CSF Env-WT (n = 939) and Env versions with single F673A (n = 1270) or double W672A/F673A (WFAA) (n = 884) mutations in the MPER. c Binding to Env on intact virions of anti-MPER bnAbs mutated in the MAPA/HCDR3 region. The intensity signals for each antibody was normalized to the WT signal after background subtraction (3R n = 1049, WT n = 1664, 3R WG n = 1437, WG n = 1444). d Correlation between potency (IC50 values for neutralization, determined by cell entry inhibition assay) and mean F/v values determined by quantitative STED in the previous samples. The statistical significance was assessed by Kruskal–Wallis test. If not noted otherwise, differences were not significant at the 0.05 level. Results are shown in box-plots (center line, median; square, mean; box, IQR; whiskers, upper and lower inner fences)