Fig. 1.
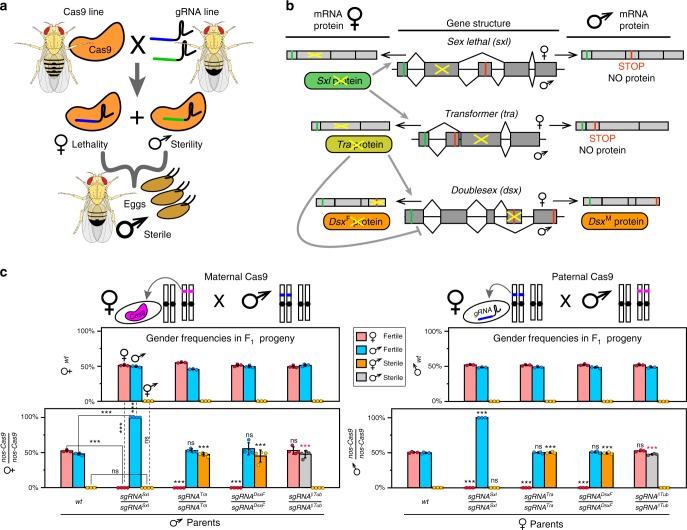
Precision guided sterile insect technique (pgSIT), an assessment of gene targets with single guide RNAs (sgRNAs). a A schematic of pgSIT utilizing two components of the binary CRISPR/Cas9 system, Cas9 and gRNAs, maintained as separated homozygous lines, their cross results in simultaneous knockouts of a gene required for female viability and a gene required for male fertility resulting in survival of only F1 sterile males. b A schematic of sex-specific alternative splicing in sxl, tra, and dsx regulated by female expression of Sxl and Tra proteins (gray lines) (modified from ref. 68). Disruption of female-specific exons of key sex-determination genes, sxl, tra, and dsx, disrupts female development. PgSIT exon targets indicated by yellow crosses. c Bar graphs of average gender frequencies in F1 progeny. Two top panels depict gender frequencies from bidirectional control crosses of homozygous sgRNA lines to wild type (wt), indicating that both fertile females and males (♀ and ♂) are present at similar ratios, but no sterile intersexes (⚥) were identified. Bottom two panels show gender frequencies from crosses of homozygous nanos-Cas9 (nos-Cas9) to wt (control) and four homozygous sgRNA lines (experiment). Independent of maternal or paternal Cas9 inheritance, 100% of trans-heterozygous sgRNASxl females were lethal, 100% of trans-heterozygous sgRNATra and sgRNADsxF females were masculinized into sterile intersexes (⚥), and 100% of trans-heterozygous sgRNAβTu males were sterile. Gender frequencies and fertility in trans-heterozygotes were compared to those in the corresponding progeny of control crosses with nos-Cas9 (solid lines) or sgRNAs (dashed lines) and wt flies. Bars represent means ± SD for three/four independent groups of parental flies. P>0.001*** by a t test assuming unequal variance (black *) or, for male sterilization by Pearson’s chi-squared test for contingency tables (red *)