FIG 4.
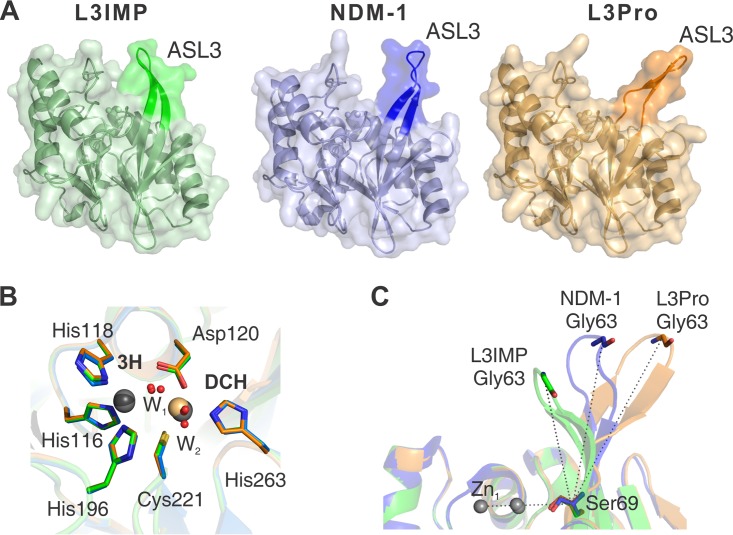
X-ray crystal structures of the L3IMP and L3Pro variants compared to NDM-1. Crystal structures are indicated by color: NDM-1 (PDB code 3SPU, chain D) in blue, L3IMP in green (PDB code 6C6I, 1.65 Å), and L3Pro in orange (PDB code 6CAC, 1.80 Å). The images were generated after the complete alignment of NDM-1 and the two L3 variants. (A) The loop L3 position is highlighted in a darker color. (B) Relevant conserved amino acids from the active sites of NDM-1 (blue), L3IMP (green), and L3Pro (orange) are represented by sticks; metal ions (Zn(II) in gray and Cd(II) in light orange) and water molecules (red) are represented as spheres. The position and orientation of the metal ligands is conserved among the three structures. The distances between the ions in the two sites are very similar (≈3.8 Å) among the different proteins. The positions of the ions in the DCH site display a slight variability among the structures, while in the 3H site the position is unchanged. (C) Angle determined by the loop L3 and the plane of the active site of each mutant. Angles were calculated between Zn1, Cα of Ser69, and Cα of Gly63. The values obtained for each loop L3 were as follows: L3IMP, 68°; NDM-1, 88°; and L3Pro, 110°.