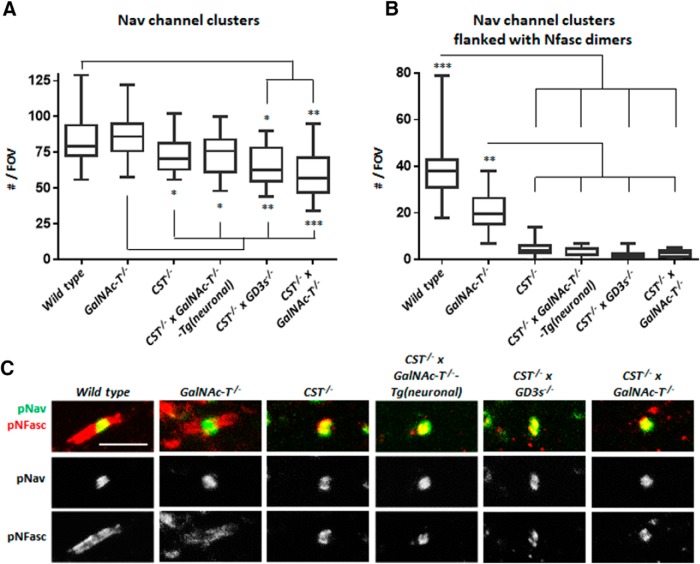
Figure 4.
Loss of both sulfatide and complex gangliosides results in a modest reduction in CNS Nav channel cluster number and sulfatide deficiency reduces NF155 presence at paranodal loops. A, The number of Nav channel clusters significantly decreases in glycolipid deficient mouse OpNs (n = 3–4/genotype). Reintroducing a- and b-series gangliosides into neurons rescues this feature. B, Nav channel clusters flanked by normal paranodal NF dimers are significantly reduced in number when sulfatide is not expressed (n = 2–3/genotype). Box-and-whisker plots are used to display the spread of all data points collected from each animal and means were calculated per genotype for statistical analysis (one-way ANOVA followed by Tukey's post hoc tests to compare multiple comparisons, indicated on the graphs as follows: *p < 0.05; **p < 0.01; ***p < 0.001). C, Representative images of OpN sections from each genotype double-immunostained for pan-Nav (pNav) antibody (green) and pNFasc antibody (red). Na channel clusters were observed in every genotype. pNFasc immunostaining formed a long band crossing the node and paranodes in WT and GalNAc-T−/− mice, suggesting labeling of the NF186 and NF155 isoforms, respectively. All of the genotypes lacking sulfatide expression had pNFasc staining restricted to the NoR and colocalizing only with pNav staining, suggesting the presence of only the NF186 isoform of NF. Scale bar, 5 μm.