Figure 2.
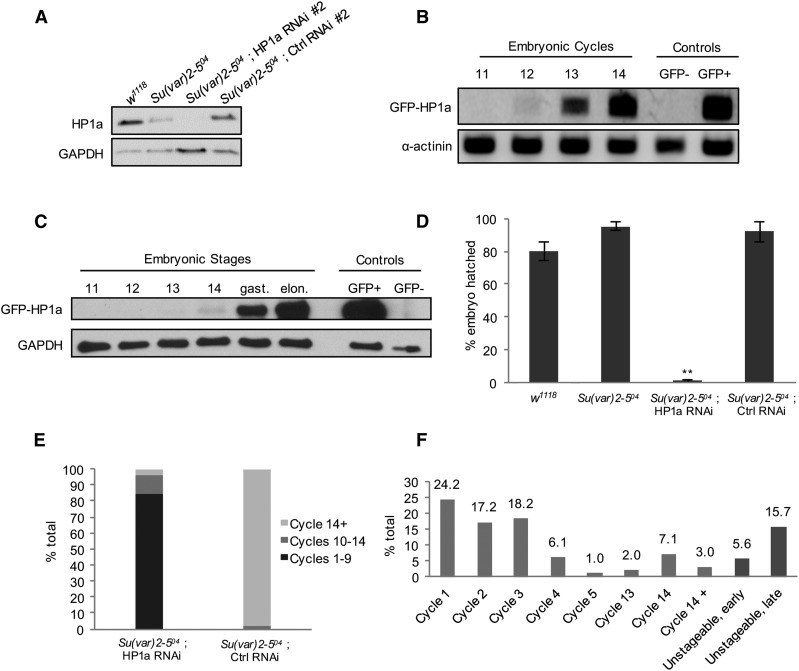
Embryos with a reduced load of maternal HP1a are developmentally delayed or arrested. (A) Embryos from HP1a-deficient females inherit a reduced amount of HP1a. HP1a expression levels in stage 14 eggs (equivalent to unfertilized embryo) collected from w1118, Su(var)2-504 heterozygote, Su(var)2-504/+ ; HP1a RNAi #2, and Su(var)2-504/+ ; control RNAi #2 females. Embryos from an HP1a RNAi female show an undetectable level of HP1a. GAPDH expression serves as a loading control. (B and C) The Zygotic transcript of HP1a is first detected at cycle 12 and protein at cycle 14. Wild-type females were crossed to EGFP-HP1a knock-in males to selectively label the paternal zygotic copy of HP1a. Embryos at different stages were collected, and probed for the presence of newly transcribed and translated paternal zygotic copies of EGFP-HP1a. (B) The GFP-HP1a transcript is first detected in embryonic cycle 12. Embryos collected from w1118 females were used as the GFP-negative control and those collected from a transgenic GFP-HP1a line (#30561; Bloomington Drosophila Stock Center) were used as the GFP+ control. The α-actinin transcript was probed as a loading control. (C) GFP-HP1a protein is first detected in embryonic cycle 14, and is expressed in an increasing manner in the later gastrulation and elongation stages. Embryos collected from w1118 females were used as the GFP-negative control and those collected from a transgenic GFP-HP1a (#30561; Bloomington Drosophila Stock Center) were used as the GFP+ control. GAPDH was probed as a loading control. (D) Embryos from females with the following genotypes, w1118, Su(var)2-504 heterozygous, HP1a RNAi (#1 and #2), and control RNAi (#1 and #2) in the Su(var)2-504 heterozygous background, were collected and the number of embryos hatched was scored after 30 hr. The average hatching rate was 80% or above in the w1118, Su(var)2-504 heterozygous, and control RNAi samples, while embryos from HP1a RNAi females did not hatch. ** P-value ≤ 0.001. n, w1118 = 382; n, Su(var)2-504 = 429; n, HP1a RNAi = 540; and n, control RNAi = 1101. Error bars indicate the mean percentage ± SEM in three biological replicates. (E) Embryos from HP1a-deficient females are developmentally delayed or arrested. The 3–5-hr-old embryos were staged and counted. In embryos from HP1a-deficient females, 85% of the embryos are in cycles 1–9, 10% in cycles 10–14, and 5% beyond cycle 14. In embryos from control females, 98% are in cycle 14 or beyond and 2% are in cycles 10–14. These values do not include the unstageable, early-, and late-stage embryos shown in (F). Embryo counts from both HP1a RNAi lines (#1 and #2) and control RNAi lines (#1 and #2) in the Su(var)2-504 heterozygous background were averaged in each biological replicate. The overall mean percentage was calculated from two biological replicates. n, HP1a RNAi = 342; n, control RNAi = 412. (F) Distribution of embryonic stages in embryos from HP1a-deficient females. Each 3–5-hr-old embryo was scored according to mitotic cycles. Of the embryos, 60% are arrested at cycles 1–3, and 10% of the embryos were found at cycle 14 or beyond. Nuclear organization in 21% of the embryos was highly disrupted and was categorized as unstageable, early (before cycle 14), or unstageable, late (cycle 14 or beyond). Embryo counts from both HP1a RNAi lines (#1 and #2) in the Su(var)2-504 heterozygous background were averaged in each biological replicate. The overall mean percentage was calculated from two biological replicates. n = 342. Ctrl, control; EGFP, enhanced GFP; RNAi, RNA interference.