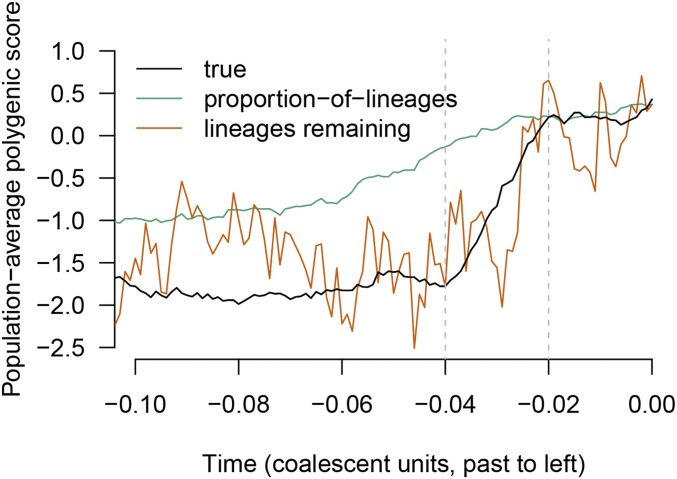
Figure 3.
The behavior of two of the proposed estimators, the proportion-of-lineages estimator and the lineages-remaining estimator, in a single simulation. The true time course of the population-average polygenic score is shown in black. The trait is under selection upward from 0.04 to 0.02 coalescent units in the past, leading to a shift of approximately two standard deviations in present-day units. Between the present and the most recent period of selection, the proportion-of-lineages estimator performs well. During the period of selection (and before), the ancestors of the sample are no longer representative of the ancestral population and the estimator becomes biased. In contrast, the lineages-remaining estimator is variable at all times but is less affected by bias associated with the period of selection. (For readability, results for the waiting-time estimator are not shown, but they are similar to the lineages-remaining estimator.) The estimates shown are formed from simulated coalescent trees for a sample of 200 chromosomes.