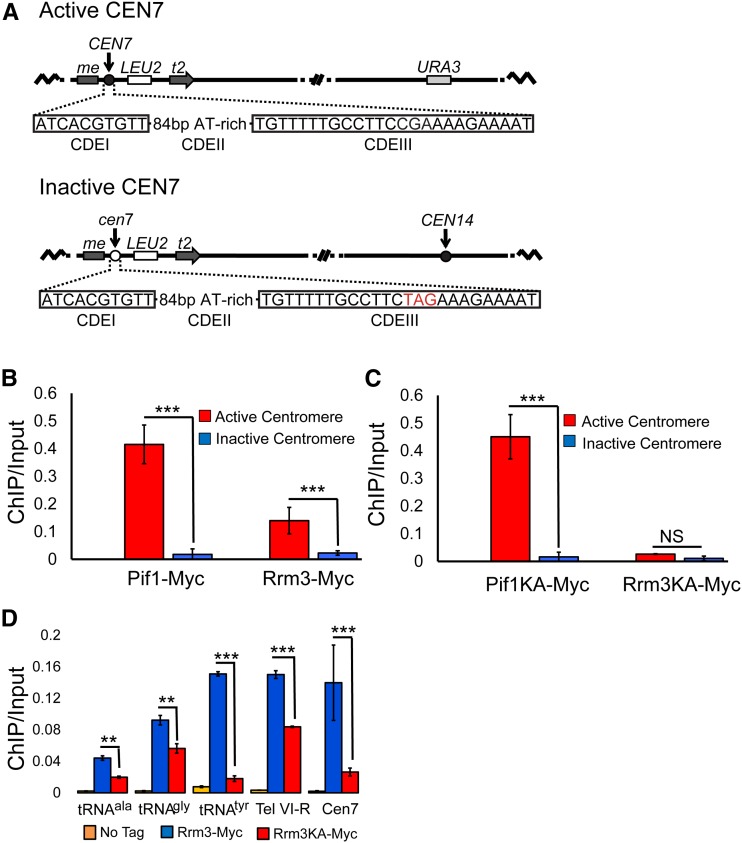
Figure 2.
ScPif1 and Rrm3 bind poorly to an inactive CEN7. (A) Diagram of the structure of active (CEN7) and inactive (cen7) centromeres, and their positions on chromosome XIV. The nucleotides in red are the changes that eliminate CEN7 function. (B) ChIP-qPCR was used to determine the levels of ScPif1 and Rrm3 binding to active CEN7 (red bars) and inactive cen7 (blue bars). (C) Helicase-inactive ScPif1 but not helicase-inactive Rrm3 binds active CEN7. The experiment is the same as in (B), except that binding of helicase-inactive proteins, ScPif1-K264A and Rrm3-K260A, was monitored. Untagged strains, which were used as negative controls, are not shown because there was no visible binding at this scale. (D) Binding of Rrm3-Myc and Rrm3-K260A-Myc to other Rrm3-sensitive sites; three tRNA sites [tDNAala (tA(AGC)F), tDNAtyr (tY(GUA)F1, and tDNAgly (tG(GCC)J2], one telomere site (Tel VI-R), and the ectopic CEN7 site from (B and C). The data for CEN7 are the same as in (C) but shown at a different scale. Blue bar indicates the binding of Rrm3. Red bar indicates the binding of Rrm3-K260A. The orange bars are the values from untagged strains. All data in (B, C, and D) are presented as [ChIP/Input] at the target sites (for historic reasons, binding was not normalized to YBL028C); error bars are one SD from the average for three independent experiments. ChIP, chromatin immunoprecipitation; NS, not significant; qPCR, quantitative PCR.