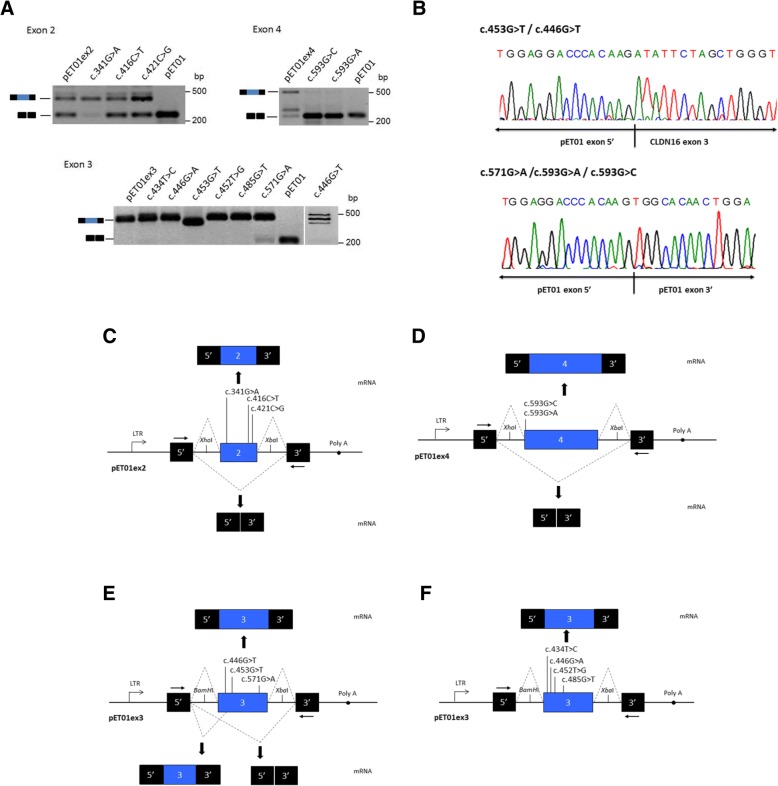
Fig. 3.
Minigene assay of CLDN16 exonic mutations revealed altered pre-mRNA splicing. a RT-PCR fragments produced by minigenes and separated by agarose gel electrophoresis. Wild-type and mutant pET01ex2, pET01ex3 and pET01ex4 constructs were transfected into COS7 cells, and the mRNAs were analysed as described in Material and Methods. All assays were performed in triplicate. The identities of the RT-PCR products are illustrated schematically on the left of each panel. b Electropherograms of anomalous RT-PCR fragments produced by minigenes containing the mutations indicated. c-f Schematic representation of minigene splicing in the presence and absence of mutations. The location of mutations is indicated. Mutations c.453G > T and c.446G > T activate a cryptic acceptor splice site internal to exon 3 that induces inclusion of a truncated exon 3. Mutation c.571G > A produced a major band corresponding to the normally spliced transcript together with a minor band that correspond to skipping of exon 3. Both c.593G > C and c.593G > A inactivate the splice site and result in exon 4 skipping