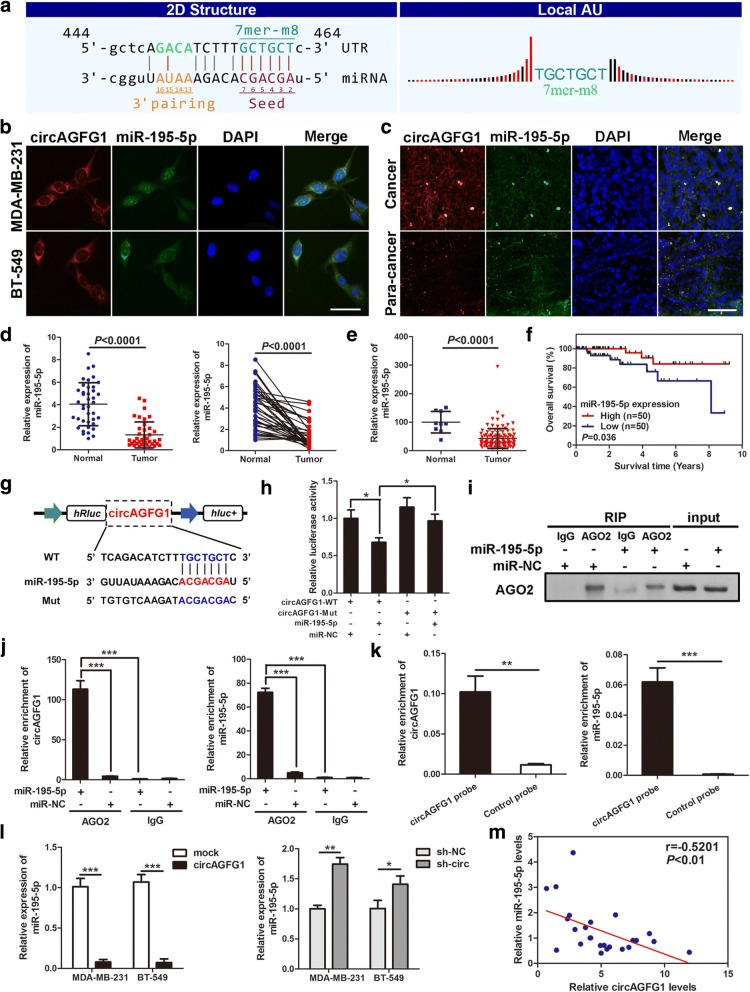
Fig. 6.
circAGFG1 functions as a sponge for miR-195-5p. a The miR-195-5p binding site on circAGFG1 predicted by targetScan and miRanda. b and c FISH was performed to observe the cellular location of circAGFG1 (red) and miR-195-5p (green) in cells (magnification, × 200, scale bar, 50 μm) and tissues (magnification, × 100, scale bar, 100 μm). d Relative expression of miR-195-5p in TNBC tissues (Tumor) and adjacent non-tumor tissues (Normal) was determined by qRT-PCR (n = 40). e Relative expression of miR-195-5p in TNBC tissues (Tumor) compared with normal tissue (normal) was analyzed using TCGA data. f Kaplan-Meier survival analysis of overall survival based on TCGA data (n = 100). g Schematic illustration of circAGFG1-WT and circAGFG1-Mut luciferase reporter vectors. h The relative luciferase activities were detected in 293 T cells after transfection with circAGFG1-WT or circAGFG1-Mut and miR-195-5p mimics or miR-NC, respectively. i and j Anti-AGO2 RIP was executed in MDA-MB-231 cells after transfection with miR-195-5p mimic or miR-NC, followed by western blot and qRT-PCR to detect AGO2 protein, circAGFG1 and miR-195-5p, respectively. k RNA pull-down was executed in MDA-MB-231 cells, followed by qRT-PCR to detect the enrichment of circAGFG1 and miR-195-5p. l The relative expression of miR-195-5p was detected by qRT-PCR after transfection with indicated vectors. m Pearson correlation analysis of circAGFG1 and miR-195-5p expression in 20 TNBC tissues. Data were indicated as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001