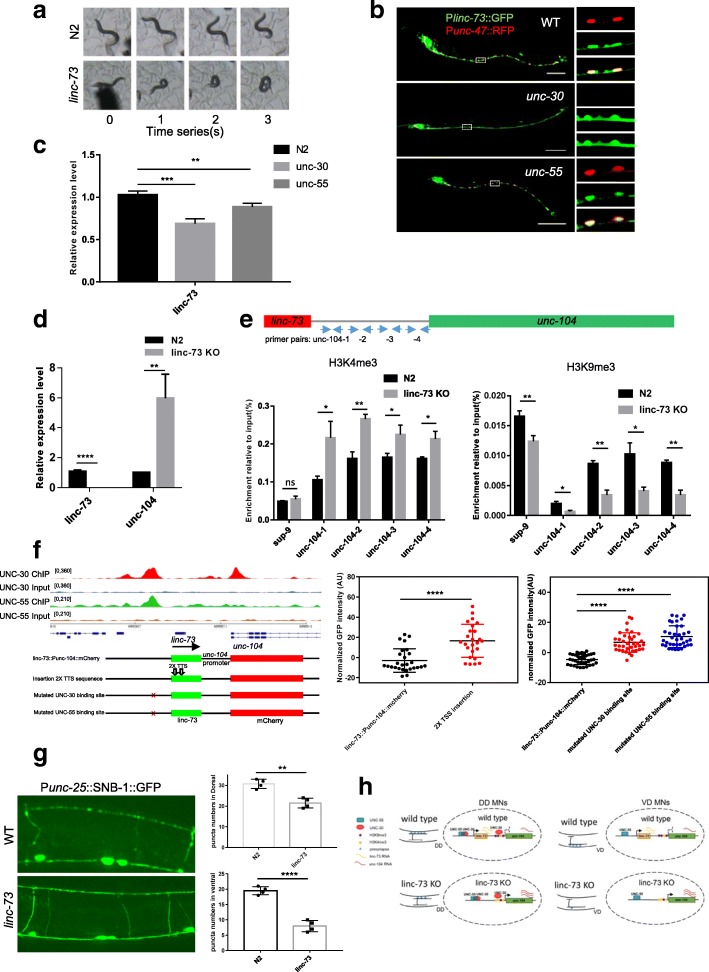
Fig. 8.
cis effect of linc-73 on the neighboring gene unc-104. a Time-lapse observation of the uncoordinated backward movement of the linc-73 CRISPR KO strain. b Expression pattern of linc-73 in the wild-type, unc-30, and unc-55 mutant backgrounds. Areas inside the dashed boxes are enlarged at the sides. Punc-47::RFP is a GABAergic marker. c qRT-PCR of linc-73 RNA levels in L2 worms of N2, unc-55(e1170), and unc-30(e191). d qRT-PCR of unc-104 mRNA levels in N2 and linc-73 KO worms (L2). e H3K4me3 (activation marker) and H3K9me3 (suppressive marker) at the promoter region of unc-104 in N2 and linc-73 mutants (L2 worms). The positions of the primer pairs used are indicated in the diagram. f Quantification of the relative expression levels of unc-104::mCherry in the cell body of D mns. Positions of the mutated UNC-30 (ΔUNC-30) & UNC-55 (ΔUNC-55) and insertion of the transcription terminal site (TTS) are shown along with the UNC-30 and UNC-55 ChIP-seq peaks. g Quantification of the dorsal and ventral presynaptic puncta (SNB-1::GFP) of DD mns in N2 and linc-73 mutants (L2). Representative images are shown. h a working model for the regulation of UNC-30 & UNC-55 on linc-73, which then regulates the expression of unc-104 to modulate C. elegans locomotion. *p < 0.05, **p < 0.01, ****p < 0.0001 by the Student’s t test. Scale bar, 50 μm