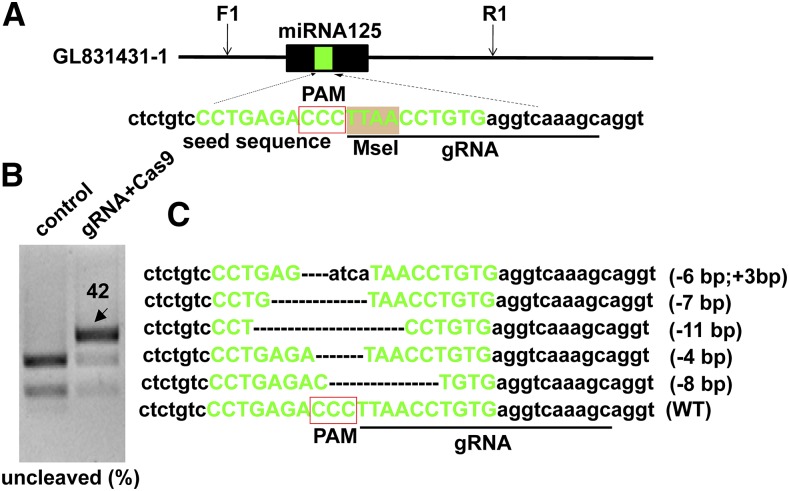
Figure 1.
Efficient disruption of miRNA125 by targeting the seed sequence using single gRNA. (A) The miRNA125 were selected as target to demonstrate the feasibility of CRISPR/Cas9 mediated mutagenesis. gRNA was designed in the seed sequence (green letters) containing a restriction enzyme cutting site (gray). The indels (insertion and deletion) were confirmed with restriction enzyme digestion and Sanger sequencing. (B) Two cleavage bands were detected in control group, while an intact DNA fragment (indicated by arrow) was observed in embryos injected with both Cas9 mRNA and target gRNA. The percentage of uncleaved band was measured by Quantity One Software. (C) Representative Sanger sequencing results from the uncleaved bands are listed. Deletions and insertions are marked by dashes and lowercase letters, and the protospacer adjacent motif (PAM) is highlighted in box. Numbers to the right of the sequences indicate the loss or gain of bases for each allele, with the number of bases inserted (+) or deleted (−) indicated in parentheses. WT, wild type.