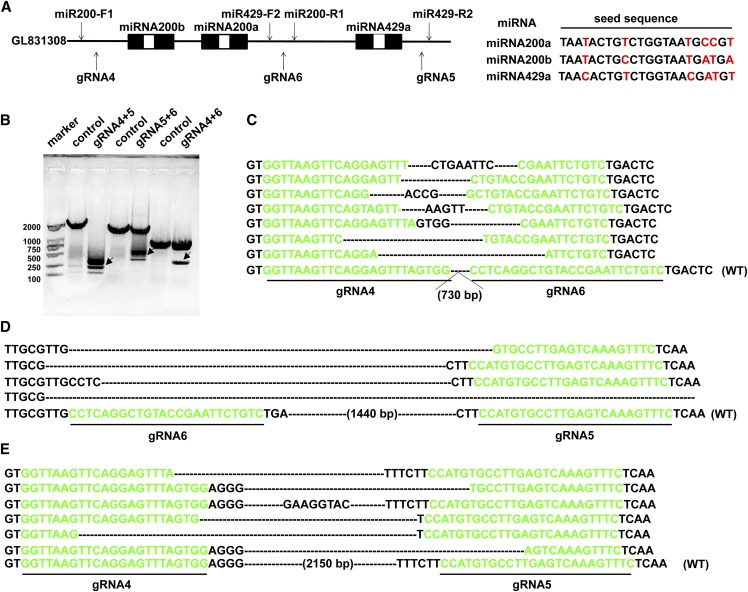
Figure 2.
Deletion of miRNA200a/200b/429a using one pairs of gRNAs and Cas9. (A) Tilapia miRNA200b, miRNA200a and miRNA429a cluster are locate on the same chromosome. They exhibit high similarity in their seed sequences. gRNA4/6 targeting miRNA200a/b (729 bp) or gRNA5/6 targeting miRNA429a (1.4 kb), or gRNA4/5 targeting the miRNA cluster (200a/200b/429a) (2.4 kb) were designed in the up and downstream of the seed sequences. (B) PCR amplification indicated successful deletion of the fragments with expected size (indicated by arrow). (C, D, E) Sequencing of the PCR products further confirmed the deletions. Deletions are marked by dashes. WT, wild type. miR200-F1/miR200-R1 and miR429-F1/miR429-R1 were designed for detection of miRNA200a/200b and miRNA429 deletions respectively. miR200-F1 and miR429-R1 were used for detection of miRNA200a/200b/429 fragment deletion.