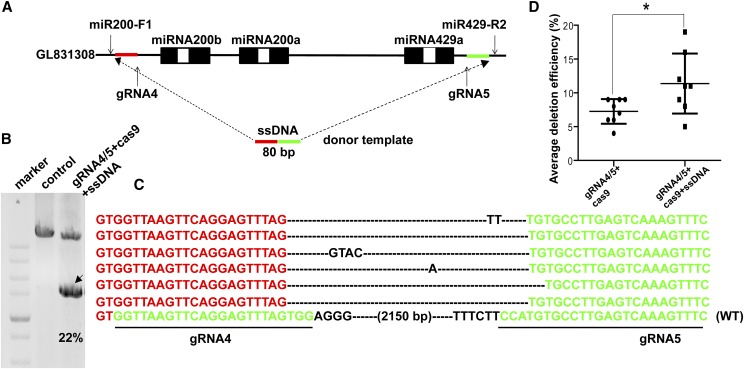
Figure 3.
Targeting efficiency was significantly increased using one pairs of gRNAs and ssDNA donor. (A) ssDNA oligonucleotide with 80 bp left and right homology arms were co-injected with two gRNAs and Cas9 mRNA into one-cell stage tilapia embryos. (B) PCR amplification showed successful deletion of the miRNA200a/b/429a fragments (indicated by arrow). miR200-F1 and miR429-R1 were used for detection of miRNA200a/200b/429a fragment deletion. (C) Sequencing of the PCR products further confirmed the deletions. gRNA sequences are underlined. Deletions are marked by dashes. WT, wild type. (D) The average deletion efficiency of eight F0 fishes (11.4%) was significantly higher than that without using ssDNA (7.2%). Data are expressed as the mean ± SD. Differences between groups were tested by independent-samples t-test. Asterisk indicates significant difference between groups (P < 0.05).