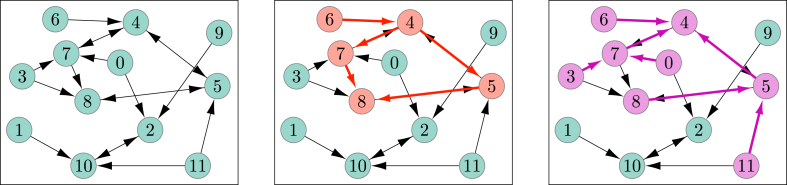
Fig. 9.
(Left) A twelve-individual population, after the a priori assignment of who would transmit to whom if ever infected by the SIR disease (the delay until transmission is not shown). Half of the nodes have zero potential infectors and half have 3. Half of the nodes have 1 potential offspring and half have 2. So the offspring distribution has PGF while the ancestor distribution has PGF . (Middle) If node 6 is initially infected, the infection will reach node 4 who will transmit to 5 and 7, and eventually infection will also reach 8 and 2 before further transmissions fail because nodes are already infected. If however, it were to start at 9, then it would reach 2, from which it would spread only to 10. (Right) By tracing backwards from an individual, we can determine which initial infections would lead to infection of that individual. For example individual 4 will become infected if and only if it is initially infected or any of 0, 3, 5, 6, 7, 8, or 11 is an initial infection.