Figure 5. Mechanistic Studies for PS-Modified DNA Oligonucleotides on Cpf1-Mediated Genome Editing.
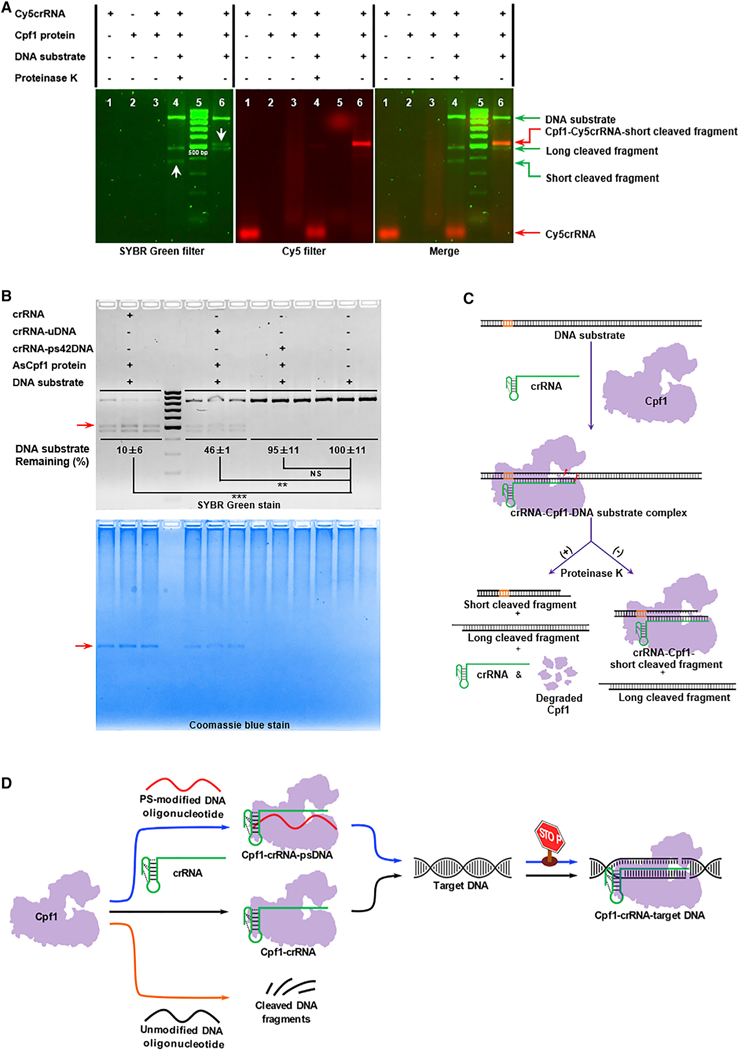
(A) Analysis of fluorescently (Cy5) labeled crRNA-mediated in vitro cleavage reactions under SYBR (left) and Cy5 (middle) filters. A merged image is shown at right. The two white arrows in the left image indicate the band shift from lane 4 (apparent size < 500 bp) in the presence of Proteinase K to that in lane 6 (apparent size > 500 bp) in the absence of Proteinase K.
(B) Top: quantification of DNA substrate bound with AsCpf1 in the presence of crRNA, crRNA-uDNA, or crRNA-ps42DNA. The uncleaved DNA substrate was quantified by densitometric analysis and normalized to the untreated DNA substrate. Data represent the normalized percentage of DNA substrate remaining (**p < 0.01 and ***p < 0.001, two-tailed t test; NS, not significant). Bottom: localization of the complex of Cpf1, crRNA, and the short cleaved DNA fragment by Coomassie blue staining. The red arrow denotes the complex composed of AsCpf1 protein, crRNA, and the short cleaved fragment.
(C) Illustration of Cpf1-mediated in vitro cleavage reactions with and without Proteinase K digestion. After the formation of crRNA-Cpf1-target DNA ternary complex, DNA substrate is cleaved. With the addition of Proteinase K, crRNA and two cleaved DNA fragments are released from the complex. Nevertheless, in the absence of Proteinase K, one DNA fragment (long cleaved fragment) without extensive interactions with crRNA and AsCpf1 protein is released from the ternary complex. The remaining ternary complex consists of AsCpf1 protein, crRNA, and the other DNA fragment (the short cleaved fragment).
(D) Proposed mechanism of inhibition for the CRISPR-Cpf1 using PS-modified DNA oligonucleotides. Normally, Cpf1 endonuclease is guided by single crRNA to recognize target DNA and induce DNA double-strand breaks (black pathway in the middle). Regarding uDNA, it has little effects on Cpf1-mediated genome editing, which may be the consequence of non-specific degradation by Cpf1 (orange pathway at the bottom). In the case of PS-modified DNA, after it interacts with Cpf1 and crRNA, the new complex cannot recognize DNA substrate, consequently inactivating genome editing (blue pathway at the top).
