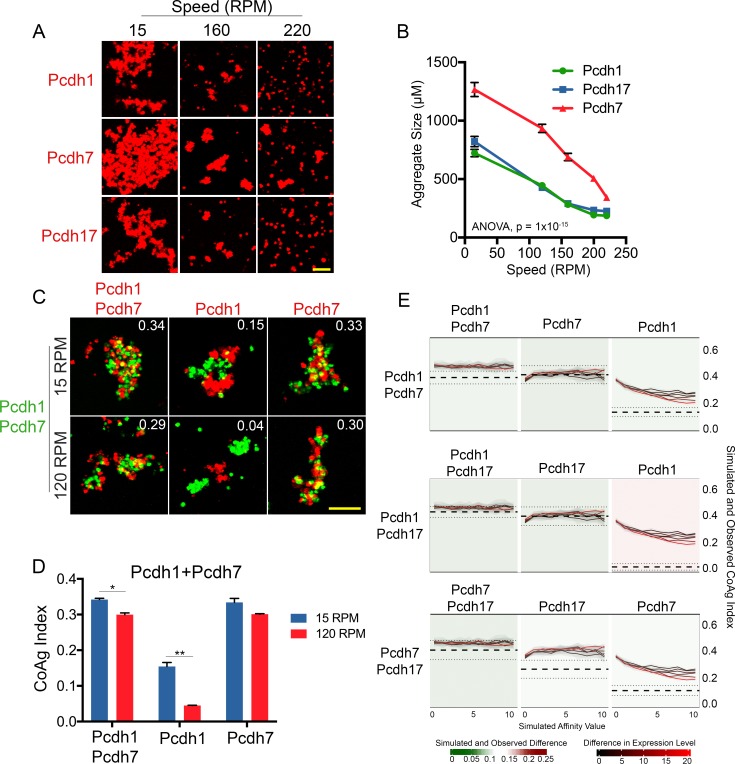
Figure 6. δ-Pcdhs possess differences in apparent adhesive affinity.
(A) Representative images of cell aggregates at select speeds. Pcdh7 cells possessed small aggregates even at 220 RPM while Pcdh1 and Pcdh17 cells dissociated. Scale bar, 100 μm. (B) Mean aggregate size at each speed. Pcdh1 and Pcdh17 were significantly different from Pcdh7 by ANOVA, p=1×10−15. Error bars indicate ±SEM. Results for each assay were determined from four independent electroporations. (C) Representative images of a mismatch coaggregation assay with Pcdh1+Pcdh7 cells. At higher speeds, Pcdh1 cells change from interfacing to segregating (middle column), while the other two populations remain intermixed. Scale bar, 100 μm. (D) Mean CoAg values of (C). Error bars indicate ±SEM, * indicates p≤0.05, ** indicates p≤0.01. Results for each assay were determined from three independent electroporations. (E) Monte Carlo simulations incorporating affinity and relative expression level capture most, but not all, mismatch assay results. We modeled the behavior of a given mismatch assay (e.g. row 1, Pcdh1+Pcdh7). The Y-axis represents the CoAg Index (simulated (solid black and red lines) and observed (thick dashed line with standard error represented by thin dashed lines). Solid lines represent simulations where the relative expression level of the two δ-Pcdhs has been varied (from 1:1 to 20:1). The X-axis represents increasing differences in apparent adhesive affinity (e.g. the left most point on the X-axis represents conditions where both δ-Pcdhs are of equal apparent adhesive affinity). In all three simulated coaggregation assays, the model predicted intermixing conditions (e.g. CoAg index above 0.2), but was not able to precisely model segregation or interfacing behaviors (compare right most graph in each row against the other two).