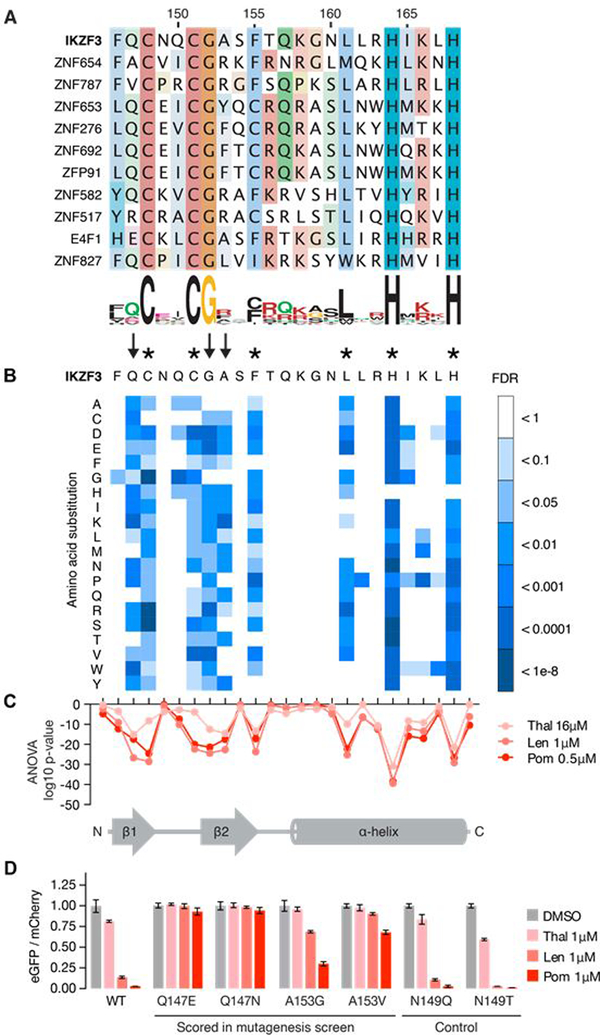
Fig. 3. Identification of amino acids required for drug-induced degradation of a C2H2 ZF degron.
(A) Sequence alignment of the 11 C2H2 ZFs with FDR<0.01 in at least one drug condition (amino acids colored by property). (B) Saturation mutagenesis screen of IKZF3 aa 130–189 in presence of lenalidomide displayed as heat map of the FDR for mutant amino acids (unpaired, one-sided t-test, FDR correction performed within each column, tech. rep. = 3). Asterisks indicate amino acids required for the ZF fold and arrows indicate non-structural IKZF3 residues required for degradation. Complete results for all 60 amino acids are located in fig. S3C (C) ANOVA p values for difference in frequency of mutant amino acids at each position in IKZF3 aa 130–189 (DMSO vs drug). Complete results for all 60 aa are located in fig. S3D. (D) HEK293T cells expressing IKZF3 ZF2 constructs in the degradation reporter were treated for 20 hours with DMSO or 1μM drug after which flow cytometry was used to measure the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 1, tech. rep. = 3, bar height is the average of tech. rep, error bars denote 95% CI).