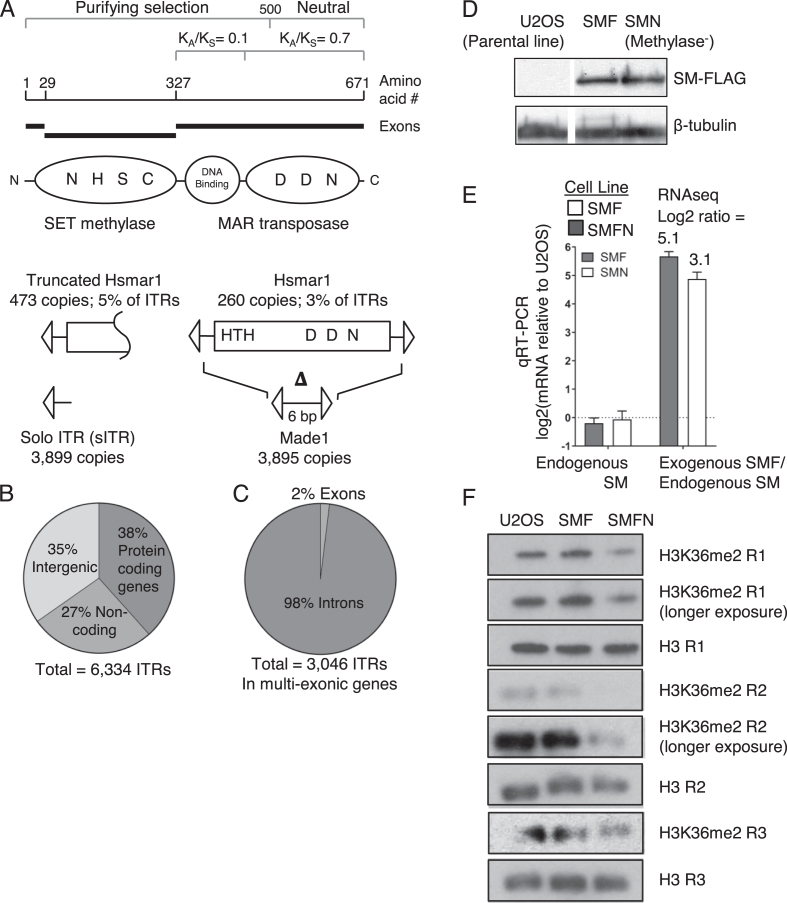
Figure 1.
Hsmar1 remnants in the human genome and expression of ectopic SETMAR. (A) The SETMAR domain and exon structure are illustrated together with the helix-turn-helix (HTH) DNA binding motifs and key active site residues in the methylase and transposase active sites. The third D residue (aspartate) that coordinates the catalytic metal ion is an N (asparagine) in SETMAR. All 260 full-length copies of Hsmar1 have inactivating point mutations and indels. Made1 elements comprise of six bp flanked by a pair of ITRs. (B and C) Distribution of the 6,334 ITRs in the human genome. (D) A Western blot for the FLAG-tagged codon optimized SETMAR in the U2OS, SMF and SMFN cell lines. SMFN has the N210A substitution of an essential residue in the SETMAR methylase domain active site. (E) qRT-PCR of the endogenous and the transgenic SETMAR in the SMF and SMFN cell lines. Figures above rightmost columns are the ratios of expression derived from the RNA-seq experiments presented in Figure 3. (F) Western blot of the indicated cell lines using an H3K36me2 antibody and a histone H3 antibody as a loading control. The lanes probed for H3 and H3K36me2, respectively, were from a single gel loaded with the same amount of protein. The same result was obtained from three biological replicates (R1 to R3) performed on different batches of cells.