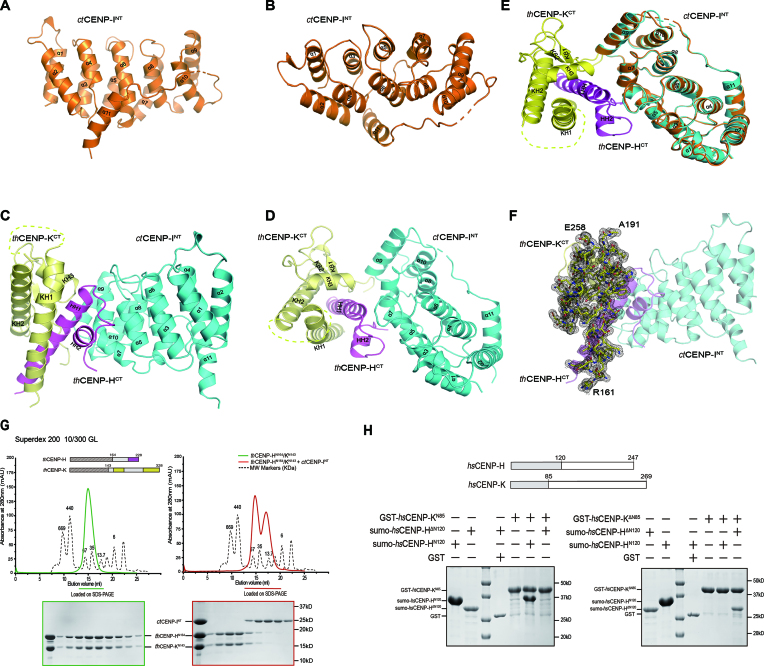
Figure 2.
Crystal structures of ctCENP-INT alone and its complex with thCENP-HCT/KCT. (A and B) Cartoon view of the crystal structure of ctCENP-INT alone in side (A) and top (B) orientations. Residues 198–205 between α10 and α11 not traceable in the electron density map are shown as dotted lines. (C and D) Cartoon view of the crystal structure of fungal CENP-H/I/K complex in side (C) and top (D) orientations. The thCENP-KCT, thCENP-HCT and ctCENP-INT are colored yellow, magenta and cyan, respectively. ( E) Electron density map for thCENP-KCT in the complex structure. The composite omit map was calculated with Phenix, contoured at 1.0σ. There is no electron density for residues 192–267 between KH1 and KH2 of thCENP-K. (F) Overlay of the ribbon diagrams of the ctCENP-INT alone (orange) and its complex with thCENP-HCT/KCT (colored yellow, magenta and cyan, respectively). (G) SEC elution profiles of thCENP-HN164/CENP-KN143 complex (left) and its mixture with ctCENP-INT (right). Peak fractions were resolved with SDS-PAGE and stained with Coomassie Blue. (H) The N-termini of hsCENP-K only bound to the N-terminal fragment of hsCENP-H (left panel), the C-termini of hsCENP-K only bound to C-terminal fragment of hsCENP-H (right panel). GST-hsCENP-KN85 and GST-hsCENP-KΔN85 were immobilized on beads and incubated with sumo-hsCENP-HN120 or sumo-hsCENP-HΔN120. The bead-bound proteins were resolved with SDS-PAGE and stained with Coomassie Blue.