Abstract
Existing methods to enrich target regions of genomic DNA based on PCR, hybridization capture, or molecular inversion probes have various drawbacks, including long experiment times and low throughput and/or enrichment quality. We developed CRISPR-Cap, a simple and scalable CRISPR-based method to enrich target regions of dsDNA, requiring only two short experimental procedures that can be completed within two hours. We used CRISPR-Cap to enrich 10 target genes 355.7-fold on average from Escherichia coli genomic DNA with a maximum on-target ratio of 81% and high enrichment uniformity. We also used CRISPR-Cap to measure gene copy numbers and detect rare alleles with frequencies as low as 1%. Finally, we enriched coding sequence regions of 20 genes from the human genome. We envision that CRISPR-Cap can be used as an alternative to other widely used target-enrichment methods, which will broaden the scope of CRISPR applications to the field of target enrichment field.
INTRODUCTION
Although next-generation sequencing (NGS) has made it possible to sequence the human genome at a cost of $1000 (1), technologies to enrich specific target DNA are required to sequence genomic regions of interest in a cost-effective manner. The most widely used methods to enrich target regions of DNA are multiplexed PCR (2,3), microdroplet PCR (4,5), target circularization using molecular inversion probes (MIPs) (6–8), and hybridization capture using nucleic acid baits (9–11). Each method has advantages. For example, PCR is easy to use, and hybridization capture can simultaneously enrich many regions. Each method also has disadvantages, such as the need for a specific device and/or multiple experimental procedures, which can take several hours to days (12–14). Typically, researchers choose a method based on their specific experimental goals by balancing several factors, which may include the robustness of base calling, coverage of the target region, uniformity of enrichment, and cost (12).
We investigated the potential of the clustered regularly interspaced short palindromic repeats (CRISPR) system as a rapid and low-cost method for target enrichment. The type II CRISPR system from Streptococcus pyogenes requires Cas9 protein (SpCas9) and two RNAs—the CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA)—to recognize and cleave both strands of the target DNA. The crRNA and tracrRNA retain their activities when fused into a single chimeric form, referred to as the single-guide RNA (sgRNA) (15). SpCas9 and sgRNA form a complex (Cas9 complex) that binds and cleaves target DNA and then remains bound to the target DNA for up to 5.5 h (16,17). We hypothesized that the Cas9 complex along with the bound and cleaved DNA (Cas9–DNA complex) could be sorted for robust target enrichment.
The CRISPR system has been utilized in various in vivo and in vitro applications. Several in vivo or in vitro applications were introduced using the wild-type CRISPR protein, catalytic dead CRISPR protein (e.g. dCas9), or various chimeric CRISPR proteins. The majority of in vivo applications of the CRISPR system are used for genome engineering (18,19). Guided nuclease (20), transcriptional regulation (21,22), epigenetic modification (23,24), and target-base editing (25,26) are well-known examples of in vivo applications of the CRISPR system. The CRISPR system has also been used to visualize target loci of genomic DNA in vivo. The CRISPRainbow (27) labels multiple genomic loci in living cells using dCas9 and modified sgRNA to recruit MS2-linked fluorescent proteins. Another method uses EGFP-tagged catalytic dead SpCas9 (SpdCas9) to visualize telomeres, DNA organization, dynamics of specific loci, and chromosomal dynamics during mitosis in living cells (28). The latter method was expanded to other CRISPR systems from Staphylococcus aureus and used to visualize multiple genomic loci simultaneously (29). Similarly, Researchers showed mRNA localization in living cells using a fluorescence-fused CRISPR protein, which was comparable to fluorescence in situ hybridization (30). Another study used a CRISPR array-acquisition system as a method to store data in living cells (31).
Several in vitro CRISPR applications have been developed. Methods for in vitro screening of off-target sites bound by sgRNAs such as GUIDE-seq (32), Digenome-seq (33), SITE-seq (34), and CIRCLE-seq (35) were developed. The CRISPR system has also been used as a molecular detection tool. Specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) was developed as a method to detect attomolar concentrations of pathogen-specific nucleic acids (36–38).
The CRISPR system has also been used for targeted sequencing. A method called engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) was developed for site-specific analysis of protein or DNA. In that method, tag-fused dCas9 and bound target DNA are formaldehyde fixed and isolated for either mass spectrometry to detect proteins interacting with the target DNA (enChIP-MS) (39) or NGS to detect physical interactions between the target DNA and genomic regions (enChIP-Seq) (40). Other applications used the CRISPR system to remove non-target DNAs from DNA libraries. For example, mitochondrial nucleic acid is one of the major non-target components of DNA sequencing libraries. Mitochondrial sequence-specific CRISPR systems were used to deplete mitochondrial DNAs from DNA sequencing libraries in RNA-seq [depletion of abundant sequences by hybridization (DASH)] (41) and ATAC-seq samples (42). In those examples, the CRISPR system was used to increase the proportion of target DNA in the DNA libraries, but not to enrich small target regions within a large genome.
Some methods use the CRISPR system to cleave and isolate target regions of DNA. Cas9-assisted targeting of chromosome segment (CATCH) (43) and CRISPR-mediated isolation of specific megabase-sized regions of the genome (CISMR) (44) are methods to analyze the sequences of large fragments of DNA. In both methods, the target region is cleaved from genomic DNA and purified by pulse field gel electrophoresis. Although CATCH and CISMR are suitable for obtaining large DNA targets, their efficiencies for enriching small DNA fragments have not been verified. Furthermore, both methods require more than a day of experiment time. Short tandem repeats (STR)-seq uses the CRISPR system to analyze short tandem repeats (45). Although STR-seq can enrich small target regions using a modified NGS flow cell, the modification of the flow cell is difficult and time consuming.
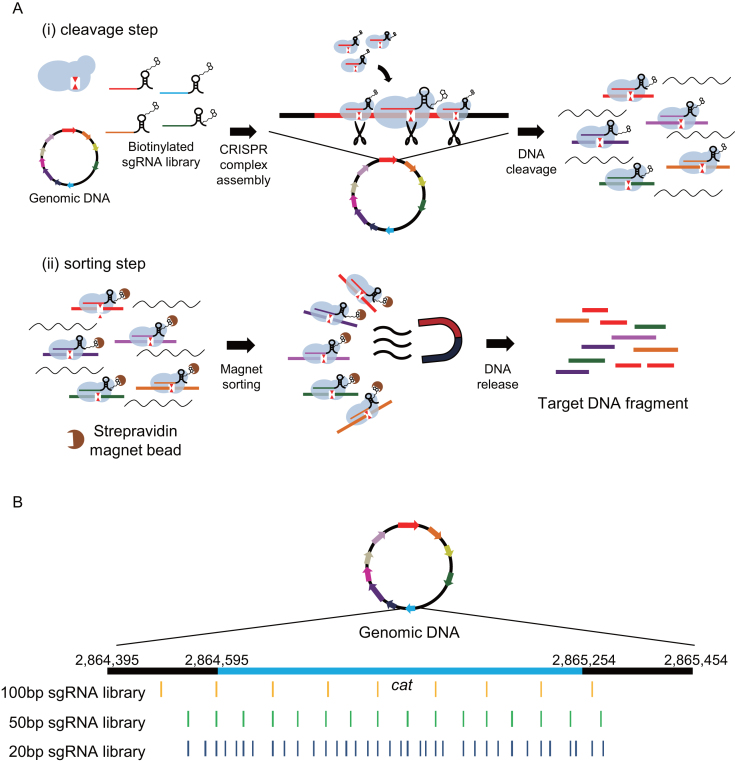
Here, we report a dsDNA target-enrichment method called CRISPR system-assisted dsDNA capture (CRISPR-Cap), which uses SpCas9 and a sgRNA library to enrich multiple small target regions from whole-genome DNA in less than 2 hours. CRISPR-Cap consists of two main steps: a target-cleavage step and a sorting step. In the target-cleavage step, multiple biotinylated sgRNAs form Cas9 complexes with SpCas9 and cleave the target genomic DNA. In the sorting step, the resulting Cas9–DNA complexes are sorted using streptavidin beads, and the cleaved target DNA fragments are subsequently released from the complexes (Figure 1A).
Figure 1.
Schematic diagram of CRISPR-Cap and sgRNA target positions. (A) Schematic diagram of the standard CRISPR-Cap procedure. In the cleavage step, SpCas9, a biotinylated sgRNA library, and genomic DNA are mixed and incubated for cleavage of target regions in the genomic DNA. In the sorting step, cleaved CRISPR-DNA complexes are bound to streptavidin magnetic beads, and target DNA is released from the complexes. (B) Three sgRNA libraries that target the same genes were designed with different cleavage densities. For example, the cleavage loci in the cat gene with each sgRNA library are represented in yellow, green and deep blue. The light blue region represents the coding sequence of the cat gene, and black regions represent 200-bp upstream and downstream of cat.
The efficiency of CRISPR-based targeting is highly variable and difficult to predict, and there is often unspecific cleavage in off-target regions. Therefore, our initial CRISPR-Cap attempts were hampered by the wide variation in CRISPR activity and the uneven cleavage rates of multiple target regions. We resolved those challenges by using a large number of sgRNAs with higher tiling of cleavage sites in the target regions. We performed enrichment with the Escherichia coli genome as a proof of concept and found that our strategy helped to improve the on-target capture ratio and the enrichment uniformity. We also found that CRISPR-Cap can be used to quantify gene copy numbers from both purified genomic DNA and whole-cell lysates. Finally, we used to CRISPR-Cap to enrich the coding sequence (CDS) regions of 20 cancer-related genes (46) in human genomic DNA. Thus, we propose CRISPR-Cap as a simple and cost-effective method for the rapid enrichment of dsDNA regions of interest.
MATERIALS AND METHODS
Cell culture and genomic DNA precipitation
We cultured E. coli EcNR2 and EcHB3 cells in Luria-Bertani (LB) media (BD Biosciences, USA) at 30°C in a shaking incubator. We harvested the cells by centrifugation and precipitated the genomic DNA using the GeneAll Exgene Cell SV Kit (GeneAll, Korea) according to the manufacturer's protocol. We purchased genomic DNA of NA12878 from the Coriell Institute (USA).
SpCas9 protein purification
Professor Hyongbum Kim's group donated the expression plasmid pET28a/Cas9-Cys, which has the SpCas9 protein-coding sequence appended with an N-terminal 6X -His tag and additional C-terminal cysteine for purification. The in vitro cleavage activity of the purified SpCas9 was previously confirmed (47). We transformed C2566 BL21-based cells (New England BioLabs, USA) with the plasmid and cultured them in LB-kanamycin (30 μg/ml) media to an optical density at 600 nm (OD600) of 0.5. We induced SpCas9 by treating the cultures with a 0.5 mM final concentration of isopropyl β-d-1-thiogalactopyranoside (IPTG) for 4 h at 30°C in a shaking incubator. We then harvested the cells and sonicated them 15 times with a 40% duty factor in 10 s bursts with a 10 s rest on ice between bursts. The lysis buffer was 20 mM Tris–HCl pH 8.0, 300 mM NaCl; 40 mL buffer/1 l cultured cells. After sonication, we centrifuged the crude extract and sonicated the supernatant overnight with Ni-NTA agarose resin (QIAGEN). We then loaded the resin-bound sample onto a column. We washed the loaded column three times with wash buffer (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 20 mM imidazole), using a buffer volume corresponding to 3× the resin volume. We subsequently eluted the protein five times with elution buffer (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 250 mM imidazole), using the same volume used for the Ni-NTA resin purification. Because the purified sample contained unwanted proteins, we filtered out small proteins using dialysis buffer [50 mM Tris–HCl pH 8.0, 200 mM KCl, 0.1 mM EDTA, 1 mM DTT, 20% glycerol, 1 tablet/50 ml cOmplete Protease Inhibitor Cocktail (Roche, USA)] and a 100 kDa pore size Amicon® Ultra Centrifugal Filter (Merck Millipore, Germany).
sgRNA library construction
To cover all the target genes or exons, we designed sgRNA libraries to include 100 bp upstream and 100 bp downstream of the target regions. To construct sgRNA libraries that target various regions in the E. coli and human genomes, we performed in vitro transcription using a DNA microarray oligonucleotide (oligo) pool (CustomArray, USA) as a template. All of the microarray oligos contained a T7 promoter sequence upstream of the sgRNA sequence. First, we amplified the microarray oligos using real-time PCR with the KAPA SYBR® FAST Bio-Rad iCycler 2X qPCR Master Mix (Kapa Biosystems, USA) until the amplification was saturated. We then amplified the products again using the KAPA HiFi HotStart Ready Mix (Kapa Biosystems) to increase the amount of template DNA. We purified the double-amplified DNA product using the MinElute PCR Purification Kit (QIAGEN) and then transcribed it using the MAXIscript® T7 in vitro Transcription Kit (Thermo Fisher Scientific, USA) according to the manufacturer's protocol. We transcribed biotinylated sgRNAs using a UTP mixture containing 80% UTP and 20% biotin-16-dUTP (Thermo Fisher Scientific). We treated the transcribed RNAs with TURBO DNase (Thermo Fisher Scientific) and purified them using an Oligo Clean & Concentrator Kit column (Zymo Research, USA). Before using them, we subjected all the sgRNA libraries to a refolding step, in which the sample was heated to 95°C and cooled at −0.1°C/s until it reached 37°C.
Cleavage step of CRISPR-Cap
We performed CRISPR-Cap in vitro. With E. coli genomic DNA, we used 1 μg genomic DNA and a 100-fold excess molar ratio of refolded-sgRNA library and SpCas9. With human genomic DNA, we used 1 μg or 100 ng NA12878 genomic DNA and a 10 000-fold excess molar ratio of refolded sgRNA library and SpCas9. We calculated the amount of sgRNA library or SpCas9 to add to the reaction as: (molecules of genomic DNA) × [number of sgRNAs in the sgRNA library (e.g. 550 with a 20-bp sgRNA library)] × (excess molar ratio) × (molecular weight of sgRNA or SpCas9)/(Avogadro's constant). We incubated the final reaction (50 μl final volume) containing genomic DNA, sgRNA library, and SpCas9 in NEB3 buffer for 1 h at 37°C in a thermocycler. After the cleavage step, we performed the sorting step.
Sorting step of CRISPR-Cap using streptavidin magnetic beads
To sort out the Cas9–DNA complexes containing the cleaved DNA, we used magnet-coated streptavidin C1 beads (Thermo Fisher Scientific). Before using them, we washed the streptavidin C1 beads three times with bead washing buffer (BWB; 5 mM Tris–HCl pH 7.5, 1 M NaCl) and then resuspended them in 50 μl 2× BWB. We mixed the washed beads directly with the product from the cleavage step and incubated the mixture for 10 min at room temperature to allow binding of the Cas9–DNA complexes to the streptavidin. We then isolated the beads using a magnetic stand and discarded the supernatant. We washed the bead pellet three times with BWB and then released the cleaved target DNA using a mixture of 50 μl nuclease-free water and 12.5 μl 0.2% SDS solution. We incubated the solution containing the released DNA for 5 min at room temperature and then purified the DNA using the MinElute PCR Purification Kit (QIAGEN) to remove any remaining SDS. Alternatively, after adding 50 μl of nuclease-free water without 0.2% SDS to the bead pellet, we could release the enriched DNA from the Cas9–DNA complexes by incubating the solution for 10 min at 65°C to inactivate the SpCas9. We used the column-purified DNA or heat-released DNA for NGS sample preparation.
Sorting step for early-release CRISPR-Cap
To perform early-release CRISPR-Cap, we mixed a 20% volume of 0.2% SDS solution directly with the product of the CRISPR-Cap cleavage step and incubated the mixture for 20 min at 37°C to promote the release of the cleaved target DNA. After SDS treatment, we purified the product using the MinElute PCR Purification Kit (QIAGEN) and used the purified DNA for NGS sample preparation.
CRISPR-Cap cleavage in a cell lysate
To perform the cleavage step in a whole-cell lysate, we harvested 1 mL saturated overnight E. coli culture by centrifugation at 16 100 rcf for 1 min. We discarded the supernatant and resuspended the pellet in 50 μl Cas9 working buffer (48). We sonicated the sample for 1 min with a 20% duty factor in 3 s bursts with 3 s rests on ice between bursts to minimize random shearing. We then mixed the cell lysate with the 20-bp sgRNA library and SpCas9. Because the amount of genomic DNA in a cell lysate is difficult to quantify, we used a 100-fold molar excess of sgRNA library and SpCas9 with 1 μg genomic DNA.
Next generation sequencing sample preparation
We prepared samples of enriched DNA for NGS with the SPARK™ DNA Sample Prep Kit for the Illumina® Sequencing Platform (Enzymatics, USA). We performed end repair and dA-tailing according to the manufacturer's protocol. We then added 10 μl NEBNext Adaptors (New England BioLabs) for Illumina to the dA-tailed DNA and performed a ligation step. We performed USER cleavage (New England BioLabs) on the adaptor-ligated sample for 15 min at 37°C and then purified the DNA using the MinElute PCR Purification Kit (QIAGEN). We performed index PCR with 16 cycles of limited PCR amplification to attach the Illumina sequencing index to the CRISPR-Cap product. We gel purified the index PCR products at a size of 250–600 bp with the MinElute Gel Extraction Kit (QIAGEN) and sequenced the purified samples using the Illumina Hiseq 4000, NextSeq, or Miseq platform.
Sequencing analysis
We used the AdapterRemoval (49) software to remove the adapter sequences from the sequencing data. We used the Burrows-Wheeler alignment (BWA) tool (50) to align the data to the reference sequence. We used SAMtools (51) with default parameters and in-house python scripts to count the sequencing depth at each position. Before aligning the adapter-trimmed sequencing data, we excluded the sgRNA template sequences that remained after in vitro transcription using an in-house python script.
Quantitative PCR
We performed quantitative PCR (qPCR) with custom primer pairs (Macrogen, Korea), the KAPA SYBR® FAST Bio-Rad iCycler 2× qPCR Master Mix, and a MyiQ Real-Time PCR machine (Bio-Rad, USA). The PCR conditions were: 3 min at 95°C, followed by 40 cycles of 20 s at 98°C, 15 s at 60°C, and 30 s at 72°C. We used E. coli EcHB3 samples as a control. We calculated fold changes in gene expression using the ΔCt value of each sample.
RESULTS
Experimental scheme for CRISPR-Cap and preliminary capture test with a single target DNA
The CRISPR-Cap procedure consists of a cleavage step and a sorting step (Figure 1A). In the cleavage step, SpCas9 is pre-complexed with biotinylated sgRNAs and used to cleave the target DNA regions. The Cas9 complex is subsequently retained at the cleaved target DNA as part of the Cas9–DNA complex. In the sorting step, the biotinylated sgRNA in the Cas9–DNA complex is bound to streptavidin-coated magnetic beads and sorted using a magnet. The cleaved target DNA is then released from the magnetic beads by a releasing agent.
First, we tried to discover the amount of Cas9 complex that was sufficient to use with the target DNA. We prepared a purified SpCas9 protein, a 545 bp linear target DNA (Supplementary Note 1), and a biotinylated sgRNA that binds to the DNA target and mediates internal cleavage by SpCas9. Polyacrylamide gel electrophoresis analysis of the purified SpCas9 protein using ImageJ (52) showed that nearly 50% of the protein in the elution product was SpCas9 (Supplementary Figure S1). Because the purity of the protein was not 100%, we conducted a test to determine the activity of the protein. We mixed various amounts of Cas9 complex (1:1 ratio of protein and sgRNA) with a fixed amount of target DNA to determine the concentration required for complete target cleavage in 1 h of reaction time (Supplementary Figure S2). We found that a 20-fold molar excess of Cas9 complex could cleave the target completely in 1 h; however, considering the different cleavage efficiencies among sgRNAs, we decided to use a 100-fold molar excess of Cas9 complex for our further experiments. We used that concentration of Cas9 complex to validate both CRISPR-Cap procedures using a single linear DNA.
After a 1 h cleavage reaction, we confirmed that the Cas9–DNA complex was retained, as previously reported (16,17) (Supplementary Figure S3, lane 2). We then used streptavidin-coated magnetic beads to sort and retrieve the Cas9–DNA complexes. We released the cleaved DNA from the sorted Cas9–DNA complexes by disrupting the complexes with 0.2% sodium dodecyl sulfate (SDS), which disrupts protein structures (53). The released DNA fragments were the size expected for the cleaved linear DNA (Supplementary Figure S3, lane 3).
To determine the most effective method for DNA release, we compared the efficacies of SDS treatment, spin-column purification, a combination of SDS treatment and spin-column purification, and RNase H digestion, which should degrade the sgRNA attached to the target DNA (54). We concluded that the serial SDS treatment and spin-column purification was the most effective method for completely releasing the cleaved target DNA from the complex (Supplementary Figure S4). We therefore used that procedure to enrich multiple genes at the genome scale.
CRISPR-Cap for multiple gene enrichment from the E. coli genome
To apply CRISPR-Cap to genomic DNA, we designed a sgRNA library to cleave 10 genes (lpd, galK, bla, prfA, cat, thyA, tolC, degS, mdh and malK) from E. coli EcNR2 (Supplementary Figure S5). We synthesized 128 sgRNAs, designed to cleave the target genes at approximately 100-bp increments, using in vitro transcription with microarray-based oligonucleotides as a template. We designated those sgRNAs as the 100-bp sgRNA library (Figure 1B, Supplementary Data 1a). Using the 100-bp sgRNA library, we performed CRISPR-Cap with purified E. coli genomic DNA. We subjected the product to typical library preparation protocols for Illumina NGS (i.e. end-repair, dA-tailing, and adapter ligation). We analyzed the sequence data using AdapterRemoval (49), BWA (50), SAMtools (51), and an in-house python program (Supplementary Note 2).
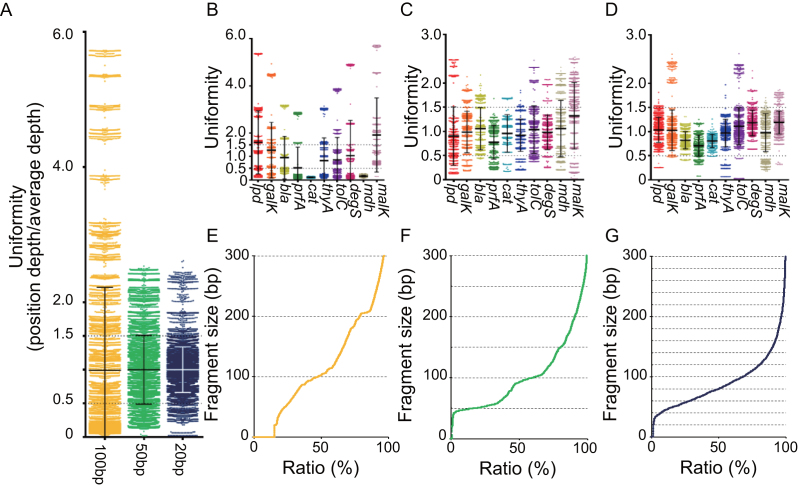
We obtained an on-target sequencing depth of 2131.0, a 41.9% on-target ratio, and 183.6 of fold-enrichment, which we calculated by dividing the percentage of on-target ratio from sequencing data over percentage of target region in chromosome {i.e. 10 584 bp (target region)/[4 647 433 bp (EcNR2 chromosome) – 10 584 bp (target region)]} (Supplementary Table S1A). However, we found that the target region was not enriched uniformly. To calculate the uniformity, we set uniformity values by dividing the sequencing depth of each target position by the average sequencing depth covering the entire target region. Using those values, we calculated the percentage of values that fell between 0.5 and 1.5 for each sgRNA library, which we called the uniform range (12). The CRISPR-Cap data for the 100-bp sgRNA library showed that 25.5% of the target region was within the uniform range, with a standard deviation (SD) of 1.23 (Figure 2A column 1). We found variation in mean uniformity values between genes such as malK and cat (Figure 2B, Supplementary Table S1b). In addition, we found partially enriched regions within single genes, such as a 300–500 bp region of prfA and a 600–800 bp region of bla (Supplementary Figure S6). Therefore, we decided to optimize CRISPR-Cap for more uniform enrichment.
Figure 2.
The uniformity of CRISPR-Cap enrichment depends on the fragment size. We evaluated the uniformity of CRISPR-Cap enrichment using the percentage of uniformity values (sequencing depth / average depth). Using the uniformity values, we calculated the percentage of uniformly enriched positions in the uniform range (0.5–1.5). (A) A scatter plot shows overall uniformity values for the on-target region with each set of CRISPR-Cap products. Black or white lines at a uniformity of 1 represent the mean value, and error bars represent the standard deviation (SD) of the uniformity value. The uniformity value of each gene in the CRISPR-Cap products cleaved at 100-bp, 50-bp and 20-bp size intervals are represented with scatter plots in (B), (C) and (D), respectively. Black bars represent the mean uniformity value of each gene, and error bars represent the SD of each gene. Size distributions of Cas9-cleaved DNA fragments using the 100-bp, 50-bp and 20-bp sgRNA libraries are represented in (E), (F) and (G), respectively. The distributions were obtained after NGS data alignment to the E. coli genome.
Optimization of CRISPR-Cap by shortening the distance between cleavage sites with higher sgRNA tiling
Although we were able to enrich 10 genes from the E. coli genome with CRISPR-Cap and the 100-bp sgRNA library, optimization was required to improve the uniformity of enrichment. We hypothesized that the uneven enrichment was caused by differences in target-cleavage efficiency among different sgRNAs (55). We assumed that a higher level of sgRNA tiling, which would cleave the target region more densely, would produce more uniform cleavage and coverage, because the low activity of some sgRNAs would be offset by higher activity of nearby sgRNAs.
We synthesized two additional sgRNA libraries that cleaved the target region into fragments of 50 and 20 bp on average, respectively (Figure 1B, Supplementary Data 1b and c). We performed CRISPR-Cap with the new sgRNA libraries and found that by cleaving the target region into smaller pieces, we could improve the on-target ratio, fold-enrichment, and uniformity. The 50-bp sgRNA library (Supplementary Table S2) and the 20-bp sgRNA library (Table 1a) produced average on-target depths of 2158.9 and 2627.7 and on-target ratios of 65.4% and 81.2%, respectively. The fold-enrichments produced by the 50-bp and 20-bp sgRNA libraries were 286.5 and 355.7, respectively, corresponding to 1.6-fold and 1.9-fold higher than that produced by the 100-bp sgRNA library.
Table 1.
Analysis of sequencing data from CRISPR-Cap enrichment with the 20-bp sgRNA library. (a) Summary statistics from target enrichment sequencing. Fold-enrichment was calculated as the on-target ratio divided by the percentage target region in the chromosome. (b) Average sequencing depth of each target gene in the on-target region.
| (a) | |||||||
|---|---|---|---|---|---|---|---|
| Target size | On-target ratio (%) | Mean depth over on target | Fold-enrichment | Breadth of coverage | |||
| 1× | 10× | 100× | 1000× | ||||
| 10 584 bp | 81.2 | 2627.7 | 355.7 | 100 | 100 | 99.8 | 97.1 |
| (b) | |||||||
| Gene | Target size | Mean on-target depth | |||||
| lpd | 1425 bp | 2718.1 | |||||
| galK | 1149 bp | 2697.2 | |||||
| bla | 861 bp | 2154.6 | |||||
| prfA | 1089 bp | 1860.8 | |||||
| cat | 660 bp | 2113.0 | |||||
| thyA | 795 bp | 2551.1 | |||||
| tolC | 1482 bp | 2914.9 | |||||
| degS | 1068 bp | 3101.4 | |||||
| mdh | 939 bp | 2573.8 | |||||
| malK | 1116 bp | 3124.1 | |||||
The 50-bp sgRNA library and the 20-bp sgRNA library improved the percentage of the target region that fell within the uniform range to 68.1% (SD = 0.51) and 90.2% (SD = 0.34), respectively (Figure 2A). Additionally, we measured the uniformity of enrichment of each target gene using the 100-bp (Figure 2B), 50-bp (Figure 2C), and 20-bp (Figure 2D) sgRNA libraries and found that cleavage into smaller pieces improved the uniformity throughout the whole target region (Supplementary Figures S7 and S8).
We also measured the size distribution of the cleaved genomic fragments by aligning the sequence data with the E. coli genome. As expected, the size of the cleavage products showed a tendency to increase in multiples of the designed fragment size of the sgRNA library (Figure 2E–G). A portion of the sequence data aligned to very short genomic regions (<100 bp), which may have lowered the quality of the Illumina-based NGS data.
Overall, we concluded that the use of a high-depth sgRNA tiling strategy to reduce the distance between cleavage sites improved the performance of CRISPR-Cap. The 20-bp sgRNA library provided maximal uniformity and on-target ratio, so we used that library for all subsequent analysis of CRISPR-Cap.
Detection of rare variants (minor allele frequency = 1%) using CRISPR-Cap
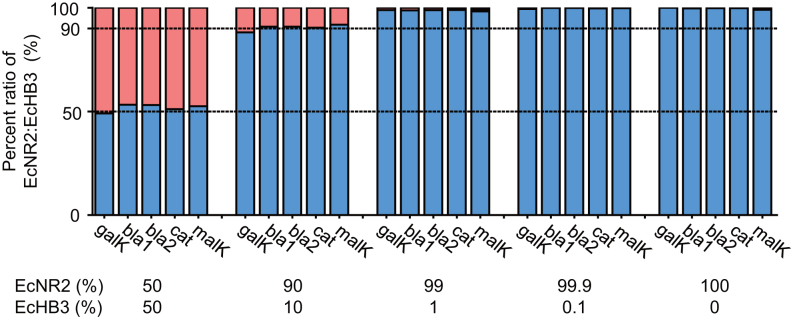
Because CRISPR-Cap enriched the target region with high uniformity, we hypothesized that the technique could be used to enrich target genes without affecting the allele ratio. We prepared a model DNA sample by mixing genomic DNA from E. coli strains EcNR2 and EcHB3. The EcHB3 strain has the same genomic DNA sequence in the 10-gene target region as the EcNR2 strain, except for five point mutations in four genes: bla, cat, galK, and malK (Supplementary Table S3). We performed CRISPR-Cap using mixed genomic DNA samples (1 μg total genomic DNA per sample) with five different mix ratios (EcNR2:EcHB3 = 50:50, 90:10, 99:1, 99.9:0.1 and 100:0). We then sequenced and analyzed the enrichment products. The sequencing data revealed that in all cases, the ratio of EcNR2 alleles to EcHB3 alleles was similar to the predefined mix ratio (r2= 0.99; Figure 3, Supplementary Table S4). CRISPR-Cap enabled the identification of the EcHB3 allele even when the allele was present at a frequency of only 1% (Supplementary Figure S9). With further optimization, we expect that CRISPR-Cap can be utilized to accurately estimate allele frequencies and detect rare mutants.
Figure 3.
CRISPR-Cap shows no significant allelic bias. E. coli EcHB3 has five different point mutations compared with E. coli EcNR2. We performed CRISPR-Cap after mixing EcNR2 and EcHB3 genomic DNA in five different ratios (EcNR2:EcHB3 = 50:50, 90:10, 99:1, 99.9:0.1, and 100:0). All five mutations were enriched with CRISPR-Cap, and the ratio of EcNR2 to EcHB3 codons from the sequencing data showed no significant allelic bias (r2= 0.99). Blue bars represent the percentage of the EcNR2 allele, and red bars represent the percentage of the EcHB3 allele (see also Supplementary Figure S9 for the enlarged plot of values at 99:1, 99.9:0.1 and 100:0).
Evaluation of gene copy number by CRISPR-Cap
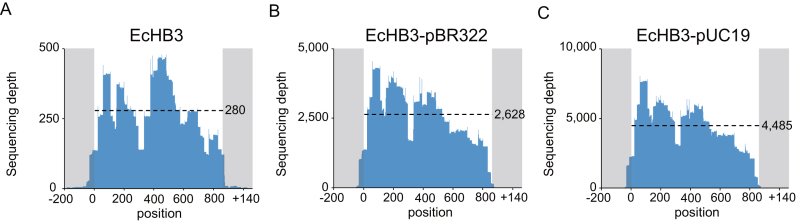
We next attempted to quantify the copy number of a gene using CRISPR-Cap. We prepared three model genomic DNA samples with different copy numbers of the bla gene, which we designated as single copy, middle copy, and high copy. We purified genomic DNA from E. coli EcHB3, E. coli EcHB3 containing pBR322 (EcHB3-pBR322), and E. coli EcHB3 containing pUC19 (EcHB3-pUC19). Both plasmids contain the bla gene when they exist in E. coli; pBR322 is a middle copy-number plasmid, and pUC19 is a high copy-number plasmid. We then performed CRISPR-Cap with the 20-bp sgRNA library and the three E. coli strains. As expected, the plasmid-containing strains showed higher sequencing depth for the bla region compared with E. coli EcHB3 (Figure 4), whereas the other target genes were enriched to similar levels in all three strains (Supplementary Table S5). The average sequencing depth for bla in E. coli EcHB3 was 280.9, which was similar to the average sequencing depth of the other target genes in that strain (306.7). The sequencing depth of bla in the EcHB3-pBR322 and EcHB3-pUC19 strains was 2628.5 and 4485.4, respectively, whereas the average sequencing depth of the other target genes in those strains was 290.6 and 293.1, respectively. By dividing the average sequencing depth of bla over that of the other target genes, we quantified the copy number of the bla gene, which were 9.0 in the EcHB3-pBR322 strain and 15.3 in the EcHB3-pUC19 strain. The increased copy numbers agreed with the average Ct value data produced by quantitative PCR (qPCR, n = 3). By setting the bla copy number in E. coli EcHB3 to one, we calculated the bla copy number in strains EcHB3-pBR322 and EcHB3-pUC19 to be 6.60 and 10.85, respectively (Supplementary Table S6). Based on the sequencing and qPCR results, we found that the average sequencing depth of the bla gene relative to the average sequencing depth of the other target genes increased as the bla copy number increased. Thus, it may be possible to use CRISPR-Cap as a method to analyze and measure changes in gene copy number.
Figure 4.
Quantification of gene copy number using CRISPR-Cap. We prepared genomic DNA samples containing three different copy numbers of the bla gene from (i) E. coli EcHB3, (ii) EcHB3 containing pBR322 (middle copy), (iii) and EcHB3 containing pUC19 (high copy). (A) E. coli EcHB3 has a single copy of bla in its genome. E. coli EcHB3 genomic DNA with either pBR322 (B) or pUC19 (C) contains middle and high copy numbers of the bla gene, respectively. Dashed lines represent the average sequencing depth of the bla gene region in each product. The average sequencing depth of the bla gene gradually increased according to the copy number.
Alternative CRISPR-Cap procedure with early release of cleaved target DNA
Following the initial characterization of CRISPR-Cap, we attempted alternative procedures to improve upon the technique and enhance its convenience. We hypothesized that the SpCas9-cleaved short DNA fragments can be differentiated from large un-cleaved genomic DNA. Based on that, we could perform NGS adapter ligation with the cleaved DNA and select the ligated DNA based on size (200–600 bp) on agarose gel during the NGS library preparation step. Thus, we could lower the cost of CRISPR-Cap by eliminating the biotin/streptavidin-based sorting step and directly releasing the cleaved DNA from the CRISPR-DNA complex prior to NGS sample preparation (Supplementary Figure S10). We named the alternative procedure ‘early-release CRISPR-Cap.’
Using the 20-bp sgRNA library, we performed early-release CRISPR-Cap and analyzed the NGS data. We obtained a mean sequencing depth of 2427.9 on the target region, indicating that the performance of early-release CRISPR-Cap was slightly less than that of the original CRISPR-Cap. The on-target ratio was still 68.9%, however, and the fold-enrichment of the target region was 301.9. Furthermore, 92.0% of the target region was located in the uniformity range with an SD of 0.48 (Supplementary Figure S11, Table S7).
Direct enrichment of target DNA from a whole cell lysate
As a second alternative CRISPR-Cap method, we tried to enrich target DNA directly from a cell lysate. We prepared whole cell lysate from E. coli EcHB3 by sonication and then added SpCas9 and the 20-bp sgRNA library to the lysate. The efficiency of CRISPR-Cap in the cell lysate was decreased compared with that of the standard CRISPR-Cap method (Supplementary Table S8). We plotted the number of sequence reads covering each position of the E. coli EcHB3 genome (Supplementary Figure S12) and found that the fold-enrichment of the target region was only 2.8, which meant that the target region was not sufficiently enriched.
Despite the low efficiency in cell lysates, we evaluated the performance of CRISPR-Cap in cell lysates for the quantification of gene copy numbers. We prepared cell lysates from the three strains used to quantify the bla copy number (i.e. E. coli EcHB3, EcHB3-pBR322 and EcHB3-pUC19). We performed CRISPR-Cap with the 20-bp sgRNA library and detected changes in the sequencing depth of the bla gene among the strains (Supplementary Figure S13). The average sequencing depth of the bla gene increased with the known copy number of bla (Supplementary Table S9). By dividing the average sequencing depth of the bla gene by the average sequencing depth of the other target genes, we calculated bla copy numbers of 16.5 in the EcHB3-pBR322 lysate and 18.4 in the EcHB3-pUC19 lysate. Therefore, although CRISPR-Cap using whole cell lysates requires further optimization, it may be feasible to use that method to detect gene copy number changes.
Applying CRISPR-Cap to human genomic DNA
We next applied CRISPR-Cap to human genomic DNA. For that, we synthesized a new sgRNA library (human 20-bp sgRNA library) capable of cleaving target CDS regions of 20 cancer-related genes into fragments 20 bp in size (46) (Supplementary Data 2c). We performed CRISPR-Cap using 1 μg NA12878 genomic DNA and a 100-fold excess of human 20-bp sgRNA library and SpCas9. The enrichment was poor, however, in terms of both the on-target ratio and the breadth of coverage (n = 3) (Supplementary Table S10). To overcome that, we tested various CRISPR-Cap conditions such as different molar ratios of genomic DNA to Cas9 complex, different sgRNA libraries, and different cleavage times.
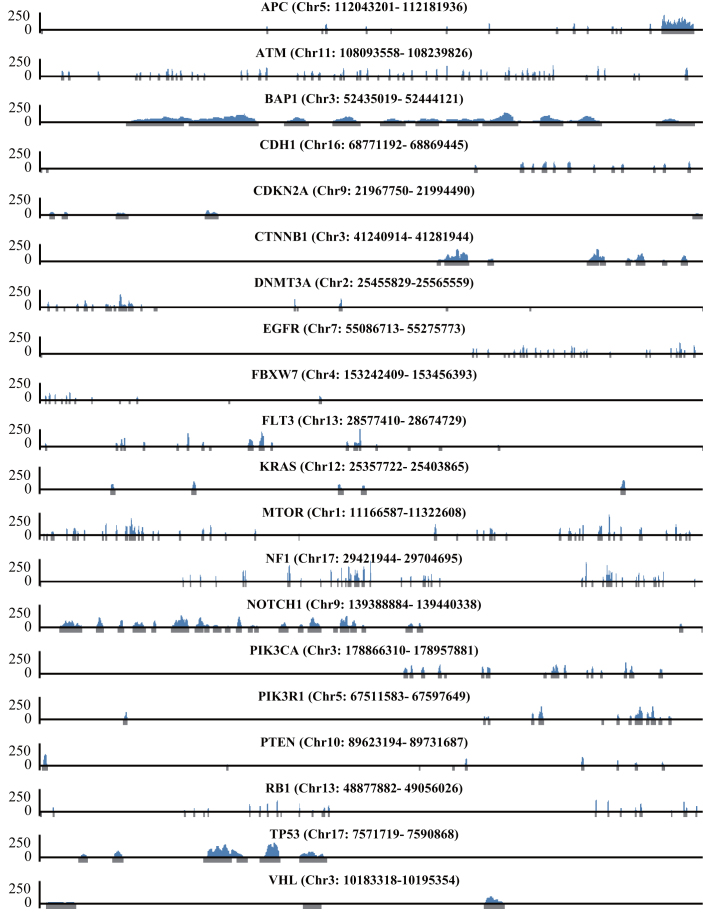
We tested different molar ratios of genomic DNA to Cas9 complex, ranging from 1:100 to 1:100 000. In contrast to the E. coli enrichment results, we found that the most promising enrichment condition was a 1:10 000 ratio of genomic DNA to Cas9 complex rather than a 1:100 ratio (Supplementary Table S10). With five additional enrichment trials using the 1:10 000 ratio, we were able to get a maximum on-target ratio of 1.24% and an average on-target ratio of 0.86%. The size ratio of the target region to the entire genome was 0.0055% (164 202 bp to 3 billion bp), so CRISPR-Cap enriched the target region by a maximum of 225.5-fold and an average of 155.7-fold. We visualized the enrichment results of one of the trials that used the 1:10 000 ratio (1:10 000_5, Supplementary Table S10) and confirmed that only the target regions were enriched (Figure 5).
Figure 5.
Enrichment of CDS regions in human genomic DNA by CRISPR-Cap. The enrichment results of NA12878 genomic DNA using CRISPR-Cap (1:10,000_5 from Supplementary Table S10). Gray regions under the x-axis indicate target regions, which include all exons of 20 genes and additional 100-bp upstream and downstream of each exon. The x-axis is the genomic position, and the y-axis is the sequencing reads count.
We next tested two different sgRNA libraries that cleaved the target region into 50-bp and 100-bp fragments, respectively. With those sgRNA libraries, we performed CRISPR-Cap with NA12878 genomic DNA and a 1:10 000 excess ratio of Cas9 complex (n = 3). Neither new library improved the CRISPR-Cap performance (Supplementary Table S11); however, we found the same tendency that we found with E. coli CRISPR-Cap: the on-target ratio increased as the size of the cleaved target fragments decreased.
Next, we tested shorter and longer cleavage durations. SpCas9 recognizes and cuts the target DNA very quickly (16). Also, the stability of the cleaved Cas9–DNA complex is known (17). We tested two alternative durations for the cleavage step: 5 min and 16 h. Despite the stability and quick cleaving ability of the Cas9–DNA complex, 1 h of cleavage time showed the best performance. Cleavage for 5 min and 16 h showed average on-target ratios of 0.062% and 0.012%, respectively (n = 6) (Supplementary Table S11). We reasoned that incubation for 5 min is not enough time for multiplexed cleavage. We speculate that 16 h of cleavage was sufficient to cleave the majority of target regions; however, we reasoned that the low enrichment efficiency was related to the lifespan of the Cas9–DNA complex, which was previously shown to be 5.5 h (17). Thus, during 16 h of cleavage time, the Cas9–DNA complex might cleave the target and become dissociated naturally.
We next examined the use of smaller amounts of genomic DNA. The initial amount of genomic DNA was 1 μg in all of the previous experiments. To find the lower limit of the initial amount of genomic DNA that could be used, we performed CRISPR-Cap starting with 100 ng or 10 ng NA12878 genomic DNA and a 10 000-fold excess of Cas9 complex composed of the human 20-bp sgRNA library and the SpCas9 protein (n = 6). The average on-target ratio of CRISPR-Cap with 100 ng and 10 ng genomic DNA was 0.65% and 0.02%, respectively, with an average fold-enrichment of 117.9 and 3.64, respectively. Also, the average percentage of the target region with 1× coverage was 79.6% and 3.03%, respectively (Supplementary Table S11). Thus, at least 100 ng initial genomic DNA was needed for CRISPR-Cap target enrichment under our conditions.
Together, the results showed that a 10 000-fold excess of Cas9 complex, high-purity genomic DNA, an sgRNA library that cleaves the target regions into small pieces, 1 h of cleavage time, and 1 μg initial genomic DNA were the best conditions for CRISPR-Cap with human genomic DNA.
Evaluating the enrichment quality of CRISPR-Cap with human genomic DNA
To examine the reliability of CRISPR-Cap with human genomic DNA, we analyzed the false-positive rate and the uniformity. To assess false positives, we compared variant calls (using GATK (56,57)) in the 1:10 000_5 data with the known genotypes of NA12878 cells (ftp://ftp-trace.ncbi.nih.gov/giab/ftp/release/). For the regions with sufficient sequencing depth and quality (≥20 × coverage and consensus quality ≥30), we observed high concordance (100%) with known heterozygous (n = 15) and homozygous (n = 9) genotypes. Those results suggest that CRISPR-Cap is applicable for the enrichment of target regions of human genomic DNA.
Next, we analyzed the uniformity of eight sequencing datasets (1:10 000_1 to 1:10 000_8, Supplementary Figure S14). We calculated the uniformity value by dividing the sequencing depth on each target position by the average sequencing depth on the sequenced target region. Although the uniformity of enrichment of the human DNA was lower than that of the E. coli DNA, 33.3–44.0% of the sequenced target region of the human DNA was within the uniformity range (0.5–1.5).
DISCUSSION
In this study, we evaluated the coverage, uniformity, and enrichment bias of the CRISPR-Cap method for the enrichment of dsDNA regions of interest. The use of higher sgRNA tiling to cleave the target region at smaller intervals greatly improved the performance of the method. CRISPR-Cap with sgRNAs that cleave the target region at 20-bp intervals showed better enrichment performance; including on-target ratio, fold-enrichment, and uniformity; than that using sgRNAs that cleave the target region at 50-bp or 100-bp intervals. Using mixtures of genomic DNA with single-nucleotide variants present at various ratios, we demonstrated that CRISPR-Cap could detect rare alleles at frequencies as low as 1%. In addition, we showed that CRISPR-Cap was able to detect differences in gene copy number in purified genomic DNA and in whole cell lysates. We demonstrated two alternative procedures that can reduce the cost of sgRNA construction and the time of genomic DNA preparation (Supplementary Figure S15). Finally, we showed that CRISPR-Cap is applicable to human genomic DNA.
In order to fully assess the feasibility of CRISPR-Cap, it is important to compare and contrast CRISPR-Cap with other target DNA enrichment methods (Table 2). A notable advantage of CRISPR-Cap is its short reaction time. CRISPR-Cap required 1 h for the cleavage reaction and 20 min for the sorting step. In preliminary experiments, only 30 min for the cleavage reaction was required for single locus targeting with one target DNA (data not shown). On the other hand, the NGS data produced from the CRISPR-Cap product had low sequencing quality. We confirmed that the relatively low sequencing quality was caused by randomly appended pseudo sequences that matched the read length of the sequencing platform (i.e. 150-bp paired-end sequencing) when the product of CRISPR-Cap DNA was shorter than the read length (58). Therefore, we used gel-based size selection after attaching the index sequence, and we processed the sequencing data with AdapterRemoval, an adapter trimming program, before proceeding to the alignment.
Table 2.
Comparison of target-enrichment methods. Comparison of technology base, main experiment steps, required time, and uniformity of enrichment product among target enrichment methods.
| CRISPR-Cap | Multiplex PCR | Microdroplet-based PCR | Molecular inversion probes | Hybridization capture | |
|---|---|---|---|---|---|
| Technology base | CRISPR system | PCR | PCR, microfluidics | molecular inversion probe-based circularization | nucleic acid hybridization |
| Experiment steps | 2 steps | 1 step | 1 step | multiple steps | multiple steps |
| Required time for experiment | <1 h | Several hours (depending on amplification cycles) | Several hours (4 h with RainDance) | 3 days | 3 days |
| Uniformity | Maximum 90.2% of TRa within uniformity rangeb | >90% within a 5-fold range of the median read depth (63) | 94.5%(4) | 60% of TRa within uniformity range (12) | 61% of TRa within uniformity range (12) |
aTR: target region.
bWith small-sized genomic DNA.
When applying CRISPR-Cap to human genomic DNA, we were able to get improved enrichment results in terms of on-target ratio, fold-enrichment, and coverage after many trials; however, even after the optimizations, the on-target ratio was still around 1% (Supplementary Table S10). To find the reason for the low on-target ratio, we analyzed the target sequences of the human 20-bp sgRNA library using Cas-OFFinder (59). Among 7118 sgRNAs, 301 sgRNAs had perfectly matched off-target sites on the genomic DNA (Supplementary Data 3). Those predicted off-target sgRNAs could cleave 0.6 million loci in the genomic DNA. Many of the potential off-target sgRNAs originally targeted intron regions, because we designed the targets to include 100 bp upstream and 100 bp downstream of the target CDS regions. Furthermore, when we changed the Cas-OFFinder parameter to allow single mismatches, the predicted number of off-target sites increased to 3.3 million loci. We aligned the eight sequencing datasets produced with the 1:10 000 ratio of DNA to Cas9 complex (1:10 000_1 to 1:10 000_8, Supplementary Table S10) to the regions starting 200 bp upstream and ending 200 bp downstream of all predicted target sites. The results showed that 6.94% and 12.95% of reads on average aligned to all perfect matched sites and single mismatches allowed sites, respectively (Supplementary Table S12). Nevertheless, the reason for the off-target results remains unclear.
DNA fragments that are erroneously produced during CRISPR-Cap and/or the genomic DNA purification step can be one of off-target source. In the eight datasets produced with the 1:10 000 ratio of human DNA to Cas9 complex, from 17.9% to 31.4% of the sequencing data was aligned to the reference genome with single-read coverage depth. To examine the distribution of the low-depth reads, we plotted the reads coverage on all the chromosomes using the 1:10 000_5 sequencing dataset (Supplementary Figure S16) and found that the low-depth, off-target coverage existed across the entire chromosomes. There was also high-depth, off-target coverage in the centromere regions, although the reason for those results remains unclear.
Next, we analyzed the error rate of the human 20-bp sgRNA libraries. We used three independently amplified sets of microarray oligonucleotides as template DNAs for in vitro transcription to produce three 20-bp sgRNA libraries, which we called human 20-bp sgRNA library batch 1, human 20-bp sgRNA library batch 2, and human 20-bp sgRNA library batch 3. Using an Illumina sequencer, we verified that 79.2%, 53.5% and 54.6% of the 20-bp target recognition spacer sequences in the template DNA of the three sgRNA libraries, respectively, were error free (Supplementary Table S13). Although we obtained the best on-target ratio with human 20-bp sgRNA library batch 2, the sgRNA library with the most errors in the 20-bp target recognition spacer regions should have produced more off-target results. Taking those results together, we concluded that the low on-target ratio for CRISPR-Cap of human genome DNA was a complex result of several factors. Nevertheless, we believe that further optimization such as off-target-free sgRNA library design and the use of error-free sgRNA library template DNA would lead to improved performance.
The dsDNA enrichment feature of CRISPR-Cap may be useful in the future. As long as a large amount of starting sample is used for CRISPR-Cap, PCR amplification during the library preparation step is not required. The enriched dsDNA can be directly attached to NGS adapters via DNA ligases. If a PCR reaction is not required, long dsDNA segments (>10,000 bp) can be captured with CRISPR-Cap for use in PacBio (60) or Nanopore-based long DNA sequencing (61,62) platforms (e.g., CATCH nanopore sequencing (43)). Currently, there are few methods available for target-capture sequencing using long-read sequencing instruments. Furthermore, PCR-free target dsDNA library preparation could be utilized for the cost-effective analysis of epigenetic marks.
In summary, CRISPR-Cap is a convenient, multiplex, target-enrichment method for use with large genomic dsDNA without further treatment. CRISPR-Cap has a short reaction time, high efficiency, and no significant enrichment bias. CRISPR-Cap can also be used to quantify gene copy numbers in small genomic DNA both from purified genomic DNA samples and from whole cell lysates. Further optimization of CRISPR-Cap may be possible for use with large genomic DNA. CRISPR-Cap is also amenable to further modifications that reduce the cost even further, such as the elimination of streptavidin magnet beads in the sorting step. We believe that further optimizations will make CRISPR-Cap useful as method to enrich target DNA in the human genome.
DATA AVAILABILITY
Raw sequencing data are available under Sequence Read Archive: SRP096854 and SRP140115.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Hyongbum Kim for his kind donation of the SpCas9 expression vector. We also thank the members of the Bang Lab for their critical comments during this work.
Authors contributions: Jeewon Lee, Ji Hyun Lee and Duhee Bang conceived of the project. Jeewon Lee and Junhyuk Cho performed all experiments. Jeewon Lee, Hyeonseob Lim, Hoon Jang, Byungjin Hwang, and Joon Ho Lee analyzed the sequencing data. Jeewon Lee and Ji Hyun Lee wrote the manuscript, and Ji Hyun Lee and Duhee Bang jointly supervised the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Mid-career Researcher Program [2015R1A2A1A1005 5972] through the National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning; Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) [NRF-2016M3A9B6948494]; Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) [NRF-2018M3A9H3024850]; Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning [NRF-2018R1A2B2001322]. Funding for open access charge: Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) [NRF-2016M3A9B6948494]
Conflict of interest statement. Duhee Bang, Jeewon Lee and Hyeonseob Lim are authors of a patent application for the method described in this paper (METHOD FOR TARGET DNA ENRICHMENT USING CRISPR SYSTEM, US.15/053,859, KR.10-2016-0022810). The remaining authors declare no competing financial interests.
REFERENCES
- 1. Park S.T., Kim J.. Trends in Next-Generation Sequencing and a New Era for Whole Genome Sequencing. Int. Neurourol. J. 2016; 20:S76–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henegariu O., Heerema N.A., Dlouhy S.R., Vance G.H., Vogt P.H.. Multiplex PCR: Critical parameters and step-by-step protocol. BioTechniques. 1997; 23:504–511. [DOI] [PubMed] [Google Scholar]
- 3. Hayden M.J., Nguyen T.M., Waterman A., Chalmers K.J.. Multiplex-ready PCR: a new method for multiplexed SSR and SNP genotyping. BMC Genomics. 2008; 9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tewhey R., Warner J.B., Nakano M., Libby B., Medkova M., David P.H., Kotsopoulos S.K., Samuels M.L., Hutchison J.B., Larson J.W. et al. . Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009; 27:1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laurie M.T., Bertout J.A., Taylor S.D., Burton J.N., Shendure J.A., Bielas J.H.. Simultaneous digital quantification and fluorescence-based size characterization of massively parallel sequencing libraries. BioTechniques. 2013; 55:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hardenbol P., Banér J., Jain M., Nilsson M., Namsaraev E.A., Karlin-Neumann G.A., Fakhrai-Rad H., Ronaghi M., Willis T.D., Landegren U. et al. . Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003; 21:673–678. [DOI] [PubMed] [Google Scholar]
- 7. Hiatt J.B., Pritchard C.C., Salipante S.J., O’Roak B.J., Shendure J.. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013; 23:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoon J.K., Ahn J., Kim H.S., Han S.M., Jang H., Lee M.G., Lee J.H., Bang D.. microDuMIP: target-enrichment technique for microarray-based duplex molecular inversion probes. Nucleic Acids Res. 2015; 43:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J. et al. . Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007; 4:903–905. [DOI] [PubMed] [Google Scholar]
- 10. Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E. et al. . Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009; 461:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cosart T., Beja-Pereira A., Chen S., Ng S.B., Shendure J., Luikart G.. Exome-wide DNA capture and next generation sequencing in domestic and wild species. BMC Genomics. 2011; 12:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J.. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010; 7:111–118. [DOI] [PubMed] [Google Scholar]
- 13. Hedges D.J., Guettouche T., Yang S., Bademci G., Diaz A., Andersen A., Hulme W.F., Linker S., Mehta A., Edwards Y.J. et al. . Comparison of three targeted enrichment strategies on the SOLiD sequencing platform. PLoS One. 2011; 6:e18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodi K., Perera A.G., Adams P.S., Bintzler D., Dewar K., Grove D.S., Kieleczawa J., Lyons R.H., Neubert T.A., Noll A.C. et al. . Comparison of commercially available target enrichment methods for next-generation sequencing. J. Biomol/ Tech.: JBT. 2013; 24:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 18. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guilinger J.P., Thompson D.B., Liu D.R.. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014; 32:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. . CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lei Y., Zhang X., Su J., Jeong M., Gundry M.C., Huang Y.H., Zhou Y., Li W., Goodell M.A.. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017; 8:16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwon D.Y., Zhao Y.T., Lamonica J.M., Zhou Z.. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun. 2017; 8:15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X.. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods. 2016; 13:1029–1035. [DOI] [PubMed] [Google Scholar]
- 27. Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T.. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016; 34:528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S. et al. . Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013; 155:1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C., Lim W.A., Huang B.. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016; 44:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelles D.A., Fang M.Y., O’Connell M.R., Xu J.L., Markmiller S.J., Doudna J.A., Yeo G.W.. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016; 165:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shipman S.L., Nivala J., Macklis J.D., Church G.M.. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017; 547:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. et al. . GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 34. Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M., Gradia S., Vidal B., Garner E., Slorach E.M. et al. . Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods. 2017; 14:600–606. [DOI] [PubMed] [Google Scholar]
- 35. Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K.. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods. 2017; 14:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. et al. . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017; 356:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F.. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018; 360:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A. et al. . Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018; 360:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujita T., Fujii H.. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 2013; 439:132–136. [DOI] [PubMed] [Google Scholar]
- 40. Fujita T., Yuno M., Suzuki Y., Sugano S., Fujii H.. Identification of physical interactions between genomic regions by enChIP-Seq. Genes Cells. 2017; 22:506–520. [DOI] [PubMed] [Google Scholar]
- 41. Gu W., Crawford E.D., O’Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L.. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016; 17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Montefiori L., Hernandez L., Zhang Z., Gilad Y., Ober C., Crawford G., Nobrega M., Jo Sakabe N.. Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Sci. Rep. 2017; 7:2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gabrieli T., Sharim H., Fridman D., Arbib N., Michaeli Y., Ebenstein Y.. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 2018; 46:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bennett-Baker P.E., Mueller J.L.. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 2017; 45:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin G., Grimes S.M., Lee H., Lau B.T., Xia L.C., Ji H.P.. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat. Commun. 2017; 8:14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yuan Y., Van Allen E.M., Omberg L., Wagle N., Amin-Mansour A., Sokolov A., Byers L.A., Xu Y., Hess K.R., Diao L. et al. . Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat. Biotechnol. 2014; 32:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H.. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014; 24:1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S., Kim D., Cho S.W., Kim J., Kim J.S.. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014; 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes. 2012; 5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Picelli S., Bjorklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R.. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014; 24:2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y., Tao W., Wen S., Li Z., Yang A., Deng Z., Sun Y.. In vitro CRISPR/Cas9 system for efficient targeted DNA editing. MBio. 2015; 10:e01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kircher M., Heyn P., Kelso J.. Addressing challenges in the production and analysis of illumina sequencing data. BMC Genomics. 2011; 12:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bae S., Park J., Kim J.S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E. et al. . Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013; 10:563–569. [DOI] [PubMed] [Google Scholar]
- 61. Quick J., Quinlan A.R., Loman N.J.. A reference bacterial genome dataset generated on the MinION™ portable single-molecule nanopore sequencer. GigaScience. 2014; 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S., Wain J., O’Grady J.. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015; 33:296–300. [DOI] [PubMed] [Google Scholar]
- 63. Hadd A.G., Houghton J., Choudhary A., Sah S., Chen L., Marko A.C., Sanford T., Buddavarapu K., Krosting J., Garmire L. et al. . Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J. Mol. Diagn.: JMD. 2013; 15:234–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available under Sequence Read Archive: SRP096854 and SRP140115.