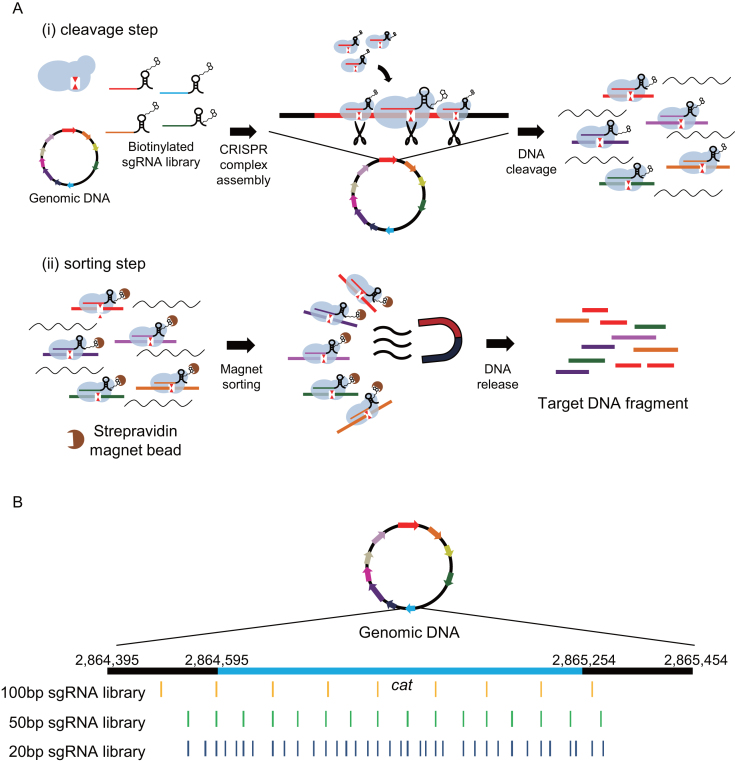
Figure 1.
Schematic diagram of CRISPR-Cap and sgRNA target positions. (A) Schematic diagram of the standard CRISPR-Cap procedure. In the cleavage step, SpCas9, a biotinylated sgRNA library, and genomic DNA are mixed and incubated for cleavage of target regions in the genomic DNA. In the sorting step, cleaved CRISPR-DNA complexes are bound to streptavidin magnetic beads, and target DNA is released from the complexes. (B) Three sgRNA libraries that target the same genes were designed with different cleavage densities. For example, the cleavage loci in the cat gene with each sgRNA library are represented in yellow, green and deep blue. The light blue region represents the coding sequence of the cat gene, and black regions represent 200-bp upstream and downstream of cat.