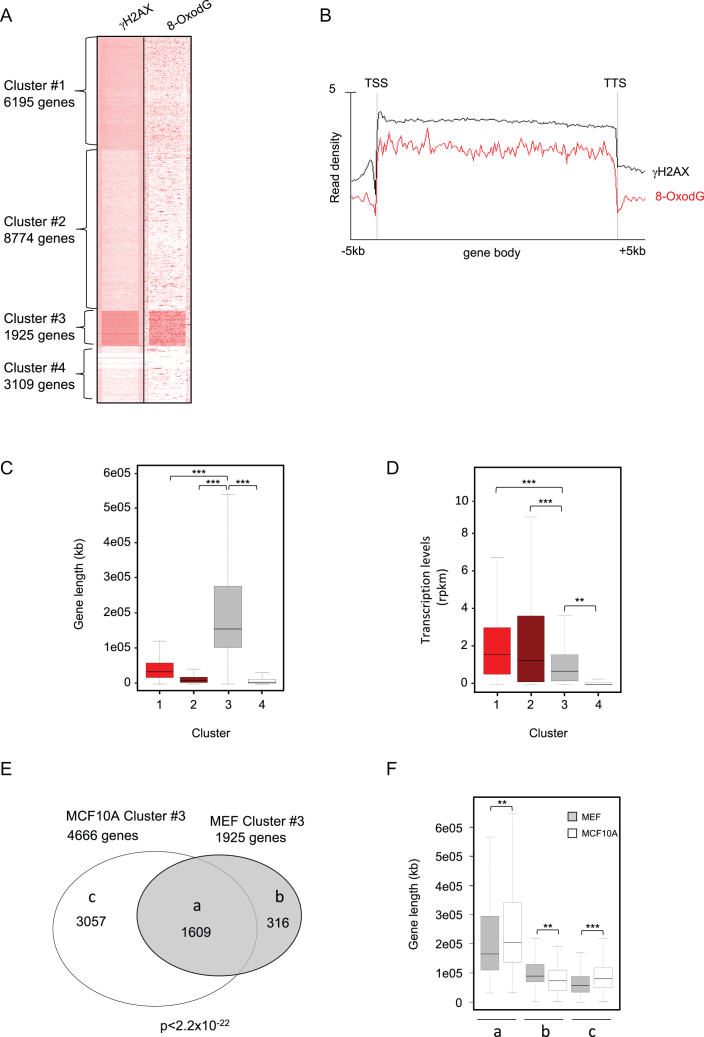
Figure 5.
(A) Heatmap showing γH2AX ChIP-Seq and 8-oxodG OxiDIP-Seq signals within the gene body, and the 5 kb both upstream the TSS and downstream the TTS of the RefSeq genes. Gene clusters identified by SeqMINER unbiased k-means clustering (Cluster #1–#4), and the number of genes contained within each cluster, as indicated. (B) Read density profiles of 8-oxodG OxiDIP-Seq (red) an γH2AX ChIP-Seq (black) in Cluster #1–#4, as indicated. (C) Box plot showing the length distribution of mouse genes within each cluster in MEFs (P < 2.2e–16, ANOVA test; ***P < 2.2e–16, **P = 2.9e–4, Bonferroni post hoc analysis for pairwise comparison). (D) Box plot showing the distribution of transcription levels (RPKM, measured by GRO-seq) of mouse genes within each cluster in MEFs (P< 2.2e–16, ANOVA test; ***P < 2.2e–16, **P = 2.9e–4, Bonferroni post hoc analysis for pairwise comparison). (E) Venn diagram showing the number of common (group a) and mouse- (MEFs, group b) or human- (MCF10A, group c) specific Cluster #3 genes; P< 2.2e–16 (Hypergeometric test). (F) Gene length distribution of the common (a), mouse-specific (b) or human-specific (c) genes in human (white bars) and mouse (grey bars), as indicated (P< 2.2e–16, ANOVA test; ***P< 2.2e–16, **P = 1.0e–5, Bonferroni post hoc analysis for pairwise comparison).