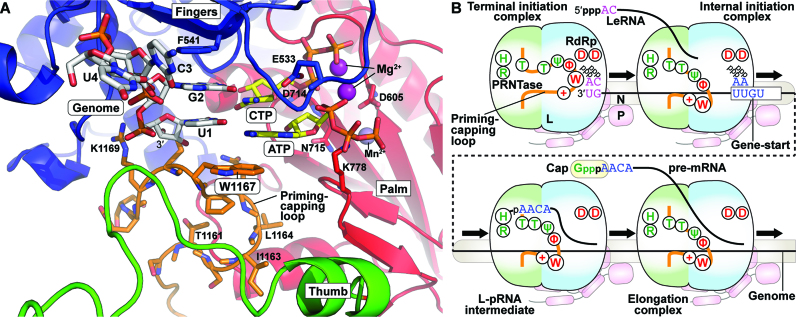
Figure 7.
The priming-capping loop of the rhabdoviral PRNTase domain governs terminal de novo initiation and pre-mRNA capping during stop-start transcription. (A) The VSV L terminal initiation complex was modeled based on the bacteriophage Φ6 initiation complex. RNA was modeled based on alignment of the core polymerases of VSV (PDB id: 5A22) and bacteriophage Φ6 (PDB id: 1HI0). The priming-capping loop (orange carbon backbone) was adjusted minimally within the cavity and the VSV L protein was energy minimized with the VSV specific RNA [genomic RNA - (3′-UGCU-5′), white carbon backbone], initial (ATP) and incoming (CTP) nucleotides (yellow carbon backbone), the two catalytic Mg2+ ions (purple) and Mn2+ (obscured) to yield the model. RdRp subdomains are individually colored: palm (red), fingers (blue) and thumb (green). Catalytic aspartates, D605 and D714, are shown on the palm. W1167 π-stacks with the initiator ATP. F541 sits stacked inline with 3′-template nucleotides, U1 and G2. (B) A model is presented for the dual roles of the priming-capping loop of the rhabdoviral L protein in terminal de novo initiation and mRNA capping. Amino acid residues in the loop required for transcription and mRNA capping are shown in red and green, respectively (see Figure 1B). The P protein is associated with the L protein and N-RNA, and essential for de novo initiation.