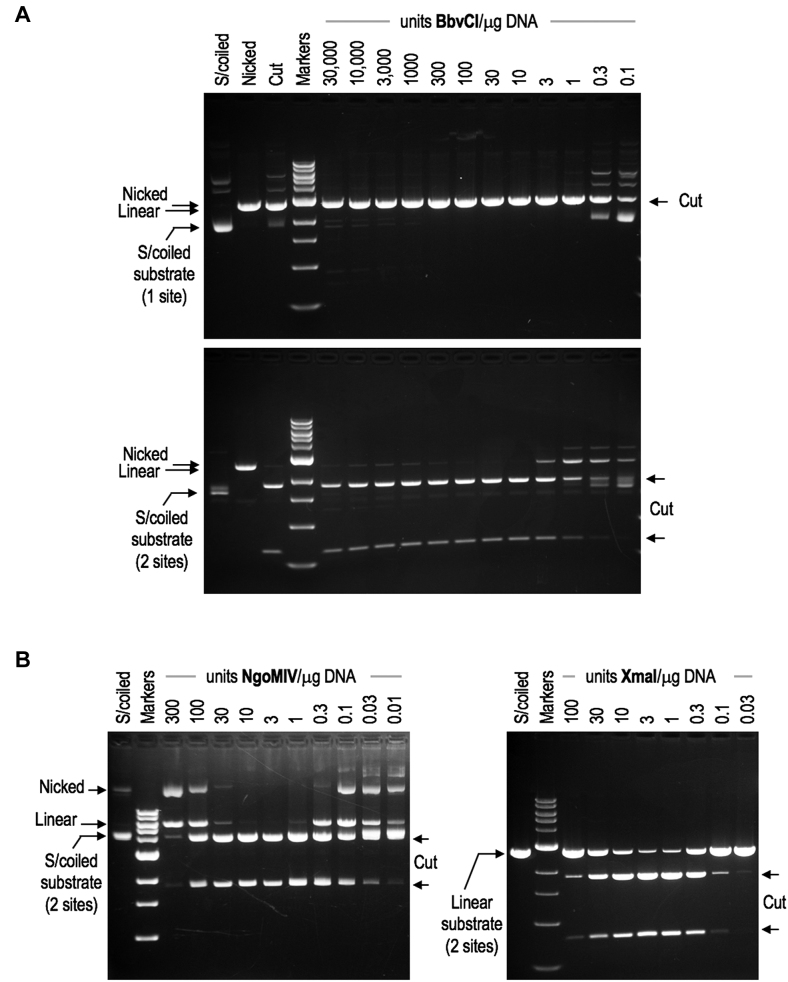
Figure 6.
Cleavage behavior of wild-type BbvCI. Panel A: Purified BbvCI was titrated in square-root 10 (3.16-fold) dilution steps on supercoiled plasmid substrates containing one target site (upper panel) or two target sites (lower panel). Each tube contained 1 μg substrate DNA ( = 0.57 pM). 280 μg of BbvCI ( = 4.4 nM of each subunit) was added to the first tube in each series, a vast excess corresponding to ∼3900 tetramers per target site (upper panel) and ∼1900 per target site (lower panel). The proportion of active molecules in the enzyme preparation is not known, and so enzyme quantities are given in units of cleavage activity, one unit being the least amount required to completely digest one microgram of substrate DNA. On this scale, a ∼30 000-fold excess of active enzyme was added to the first tube of each series. Following incubation, reactions were extracted with phenol/dichloromethane to remove DNA-bound protein, and analyzed by 1% agarose gel electrophoresis. From these and similar titrations (not shown), it is evident that 1) BbvCI cleaves substrates with one site at least as rapidly as those with two sites; and, 2) cleavage is not suppressed at high enzyme concentrations, but continues to proceed to completion. These properties are characteristic of REases that cleave efficiently when bound to only one target sequence. Panel B: Cleavage behavior of NgoMIV and XmaI. Purified NgoMIV (left panel) and XmaI (right panel) were titrated on supercoiled (left) and linearized (right) plasmid substrates each containing two target sites. Excess enzyme was added to the first tube of each titration series and serially diluted in square-root 10 steps. Following incubation, reactions were extracted and analyzed by 1% agarose gel electrophoresis. In contrast to BbvCI, cleavage by NgoMIV and XmaI is noticeably suppressed at high enzyme concentrations, and no longer proceeds to completion. This behavior is characteristic of REases that must bind two target sites at once in order to cleave, and is a property of many that act as tetramers.