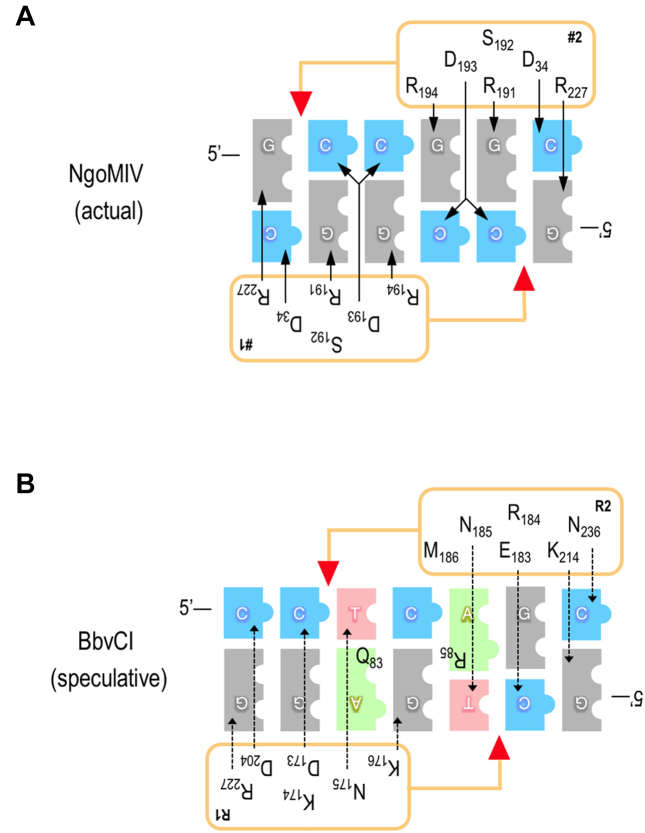
Figure 9.
Observed and proposed amino acid-base pair interaction schematics for NgoMIV and BbvCI. Panel A: The NgoMIV interactions (upper panel) are inferred from the protein–DNA co-crystal structure (26), and mirror those of the related enzymes, SgrAI and Bse634I (62,70). Panel B:The BbvCI interactions (lower panel) are speculative, and derive from aa sequence and structure alignments. The bases of the target sequences are shown as blue (cytosine), grey (guanine), red (thymine), and green (adenine) blocks. Protein subunits are depicted as rectangles alongside the DNA; red triangles indicate the cleavage positions of the catalytic sites. Amino acids inferred or speculated to determine sequence-specificity are listed within the rectangles. These are the same in the two NgoMIV subunits (labeled #1 and #2) because the subunits are identical, but they are different in the two BbvCI subunits (labeled R1 and R2) because these differ. Arrows connect amino acids to the base(s) they specify, and imply the presence of one or more hydrogen bonds. Solid arrows signify confidence in the interactions of NgoMIV. Dotted arrows signify uncertainty in the interactions of BbvCI, and those shown depict only one of several possibilities.