Figure 1.
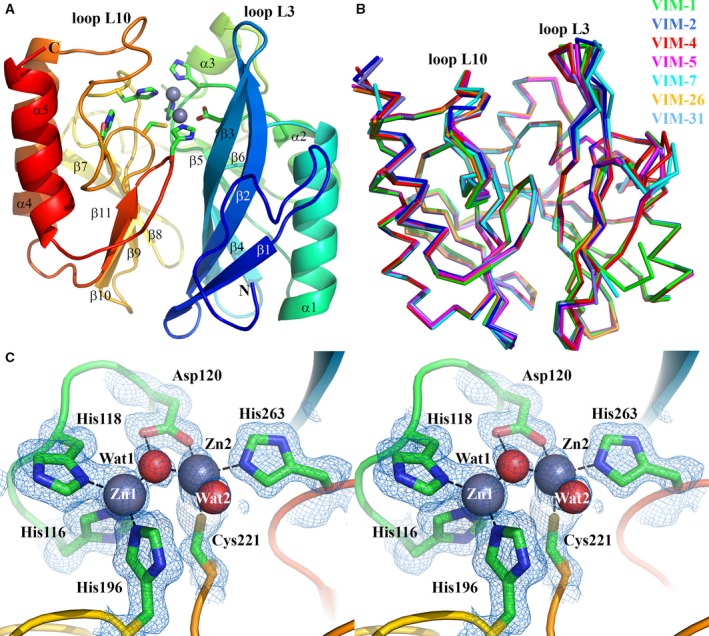
Crystal structure of VIM‐1. (A) View of the crystal structure of di‐zinc VIM‐1 showing the overall fold and location of active site. Protein main chain is colour‐ramped from blue (N terminus) to red (C terminus). Active site residues are rendered as sticks; zinc ions as grey spheres. Secondary structure elements are labelled. (B) Superposition of di‐zinc VIM‐1 with structures of other VIM enzymes (VIM‐2, pdb 4BZ3; VIM‐4, pdb 2WHG; VIM‐5, pdb 5A87; VIM‐7, pdb 4D1T; VIM‐26, pdb 4UWO; VIM‐31, pdb 4FR7; coloured as shown). (C) Stereoview of di‐zinc VIM‐1 active site. Electron density shown is 2|F o| − |F c| Φcalc contoured at 1.5 σ. This Figure was created using pymol (www.pymol.org).
