Abstract
The plant immune system is comprised of a complex network of signaling processes, regulated not only by classically-defined immune components (e.g., resistance (R) genes), but also by a suite of developmental, environmental, abiotic, and biotic-associated factors. In total, it is the sum of these interactions – the connectivity to a seemingly endless array of environments – that ensure proper activation, and control, of a system that is responsible for cell surveillance and response to threats presented by invading pests and pathogens. Over the past decade, the field of plant pathology has witnessed the discovery of numerous points of convergence between immunity, growth, and development, as well as overlap with seemingly disparate processes such as those that underpin plant response to changes in the environment. Towards defining how immune signaling is regulated, recent studies have focused on dissecting the mechanisms that underpin receptor-ligand interactions, phospho-regulation of signaling cascades, and the modulation of host gene expression during infection. As one of the major regulators of these immune signaling cascades, the plant cytoskeleton is the stage from which immune associated processes are mobilized and oriented, and in this role, it controls the movement of the organelles, proteins, and chemical signals that support plant defense signaling. In short, the cytoskeleton is the battlefield from which pathogens and plants volley virulence and resistance, transforming resistance to susceptibility. Herein, we discuss the role of the eukaryotic cytoskeleton as a platform for the function of the plant immune system.
Keywords: Actin cytoskeleton, immunity, ETI, PTI, PAMP, effector, virulence, cytoskeleton, microtubules, myosin, resistance, susceptibility, pathogen
Plant immunity
The primary function of the plant immune system is to restrict pathogen invasion and multiplication, thereby inhibiting disease and death. At the same time, the immune system must also be regulated such that beneficial interactions are not negatively impacted (Toth and Stacey, 2015), as well as permit plant growth and development (Huot et al., 2014). In both cases, the immune system plays a key role in how plants respond to the environment. Research in the area of plant-pathogen interactions has led to a model which describes two primary nodes of the immune system (Chisholm et al., 2006). In short, these separate, yet coordinately regulated pathways, are defined in large part by the source and amplitude of the immune-eliciting signal (Jones and Dangl, 2006). The first, referred to as pathogen-associated molecular pattern (PAMP; e.g., flagellin, chitin, LPS)-triggered immunity (PTI), is characterized by a rapid signaling response activated following the perception of conserved pathogen molecules by host-derived pattern recognition receptors (PRR) (Tang et al., 2017; Peng et al., 2018). In short, PTI results in the rapid activation of immune associated signaling processes, including the generation of reactive oxygen species (ROS), the induction of second messenger signaling (e.g., Ca2+), and changes in gene expression (Li et al., 2016).
As a counter response to the activation of PTI, many pathogens of plants deliver effector proteins into host cells to interfere with, or block, this initial immune signaling process (Buttner, 2016; Lo Presti and Kahmann, 2017). This, in turn, can lead to the induction of an enhanced immune signaling response referred to as effector triggered immunity (ETI). This node of immunity is mediated by the activity of nucleotide-binding leucine-rich repeat (NB-LRR) R-proteins which are activated following the recognition of delivered pathogen avirulence effectors (Su et al., 2018). Converse to PTI, ETI results in a sustained immune response, which typically manifests in the induction of the hypersensitive response (HR) – programed cell death (PCD) – which is hypothesized to function in the restriction of pathogen growth (Huysmans et al., 2017).
The cytoskeleton as a molecular and cellular scaffold of plant immunity
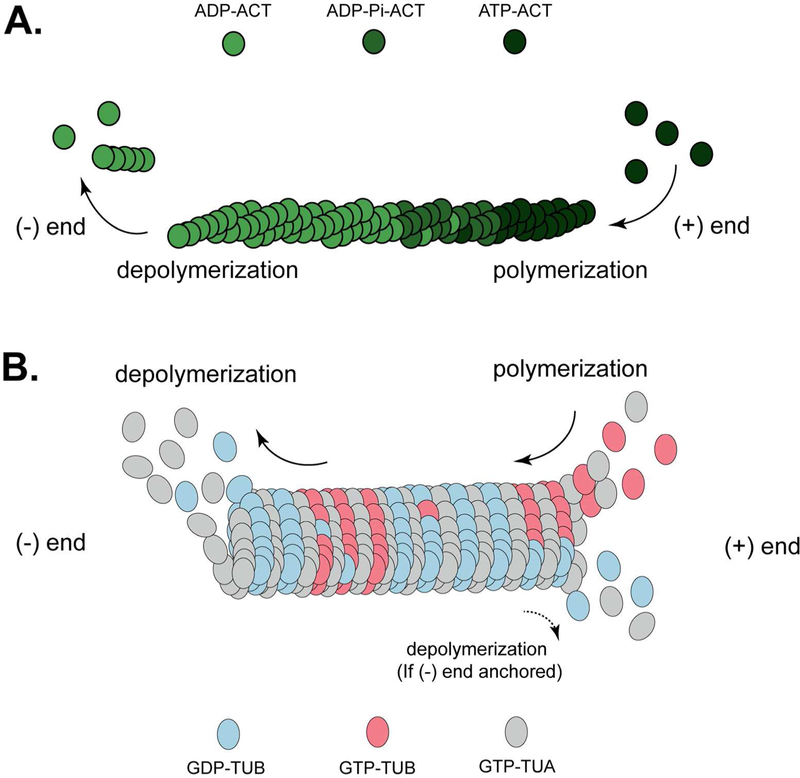
Two major classes of the plant cytoskeletal network are found in higher eukaryotes (Figure 1). The first, microfilaments (MF), commonly referred to as the actin cytoskeleton, are formed by the polymerization of globular (G)-actin into filamentous (F)-actin, a process in plants that requires the function of more than 75 actin binding proteins (Figure 1A) (Day et al., 2011). Actin is responsible for functions ranging from cytoplasmic streaming (e.g., movement of organelles) and cell division, to trafficking and endocytosis. The second, microtubules (MT), are comprised of a complex array of α/β-tubulin heterodimers, a network that is typically associated with cell growth and long-distance intercellular movement and communication (Figure 1B) (Brandizzi and Wasteneys, 2013). Both MF and MT exhibit a remarkable degree of rapid, seemingly random yet highly specific, dynamism, represented by tremendous rates of polymerization and depolymerization. Together, these patterns of cytoskeletal organization yield a highly dynamic and tightly regulated framework that connects the intercellular components of the cell to an endless suite of microenvironments and physiological processes. The eukaryotic cytoskeleton engages a variety of signaling events, including those associated with cell division and development, organelle movement, vesicle trafficking, and immunity (Porter and Day, 2015; Elliott and Shaw, 2018). As a function of the plant immune system, an abundance of data supports roles for the cytoskeleton in at least three key aspects of the immune response: 1) establishment and maintenance of signaling-competent microenvironments; 2) cellular trafficking (organelle, proteins, and small molecules); and 3) transcriptional regulation. Below, we highlight current research in each of these areas, discussing the role of each of these in immunity and the function of each as linkages between immune signaling and the dynamism of the host cytoskeleton.
Fig. 1.
Schematic diagram of cytoskeleton polymerization/depolymerization (treadmilling). A, Microfilament treadmilling. G-actin is dynamically polymerized onto the growing F-actin strand. The (+) end is defined as the site where polymerization dominates, and the (−) end as where depolymerization dominates. Actin polymerization is achieved through loading ATP-associated G-actin to the end of F-actin, while depolymerization occurs through destabilization of ADP-associated actin. B, Microtubule treadmilling. The (+) end is defined as the site where polymerization dominates, while on the (−) end, tubulin is relatively stable with dominant depolymerization. In some cases, when the (−) end is anchored, the (+) end can also have stochastically dominant polymerization. α-tubulin (TUA) and β-tubulin (TUB) form a heterodimer as the basic unit of polymerized microtubule. TUA is constitutively bound to GTP; TUB binds the growing MT filament as GTP-bound monomers, and tends to disassociate from the filament when bound to GDP.
In a typical plant cell, the vast majority of the cytoskeleton stretches from the cytosol to attachment points at or near the plasma membrane (PM). This is significant, as the PM is regarded as one of the key signaling interfaces between the host and pathogen, supporting the function of two primary classes of immune receptors: PRR complexes and the coiled-coil type NB-LRR (CC-NLR) resistance proteins. Thus, it is not surprising that the cytoskeleton-PM interface is also a key component of the signaling processes associated with receptor activation, mobilization, and signaling transduction. Indeed, as a scaffold for many of these PM-associated processes, recent work has revealed that the plant cytoskeleton selectively interacts with – either directly or indirectly – numerous membrane-localized receptors associated with immunity and signaling plant defense.
In Arabidopsis, the PM localized PRRs FLS2 (flagellin receptor) and BRI1 (brassinosteroid receptor), interact with BIK1 to form a co-receptor complex to initiate downstream signaling (Couto and Zipfel, 2017). Following ligand binding, activated PRR complexes aggregate into distinct nanodomains within the PM, where they function in immune signaling activation (Keinath et al., 2010). Indeed, a recent study demonstrates that FLS2-BIK1 and BRI1-BIK1 complexes localize in distinct nanodomains within the PM, where they further associate with different proteins required for downstream signaling (Bucherl et al., 2017). In the case of BRI1-BIK1, the nanodomain has been shown specifically interact with the MT network. This finding is significant as it provides experimental evidence that plant receptor kinases, including immune receptors, form functional complexes with the plant cytoskeleton to activate downstream signaling associated with immunity. In an additional study, it was further demonstrated that disruption of actin filament organization leads to the generation of a relatively enhanced ROS burst response following flg22 perception by FLS2 (Sun et al., 2018). In total, these studies were among the first to provide evidence supporting the hypothesis that cytoskeletal organization – and the physical interactions between PRR complexes and actin – are required for maintenance of appropriate levels of immune activation and signaling.
While conclusive data demonstrating that the plant cytoskeleton directly interacts with individual immune receptors is lacking, an abundance of data in mammalian systems does exist. For example, the nucleotide-binding oligomerization domain protein 1 (NOD1), the PRR responsible for perception of p G-d-glutamyl-meso-diaminopimelic acid (iE-DAP), requires F-actin for proper PM localization. Further, the interaction(s) between NOD1 and actin serves as an immune interface which influences actin-remodeling and control of downstream signaling (Kufer et al., 2008), including the phospho-dependent activation of the actin depolymerizing factor cofilin (Bielig et al., 2014). Similar to the activation of NOD1, the mammalian muramyl dipeptide receptor NOD2 is also recruited to the PM through its association with actin (Legrand-Poels et al., 2007). Using a pharmacological-based approach, it was demonstrated that following application of cytochalasin-D, an inhibitor of actin polymerization, both NOD1 and NOD2 signaling are activated, providing strong support for the hypothesis that the actin remodeling (including depolymerization) of PM-associated F-actin is likely a physical trigger of NOD1/2 signaling. Taken together, data from both plant and animal systems support the hypothesis that the cytoskeleton provides the necessary microenvironment to sustain the functionality of immune receptor complexes, and based on this, we hypothesize that the actin cytoskeleton is a guardee of PM-localized PRRs.
The actin cytoskeleton is required for turnover of PM-localized PRRs
During both PTI and ETI, the turnover of activated signaling complexes is mediated by receptor endocytosis, a process that functions not only to protect the plant from constitutive activation of defenses (i.e., autoimmune response), but also to support the surveillance function of the immune system (He et al., 2017). In the case of the PTI, recycling of PM-associated immune components is controlled in large part by clathrin-mediated endocytosis (CME), a process that requires the function of the actin cytoskeleton (Nagawa et al., 2012). Well-studied in animal and yeast models, CME is initiated by loading the clathrin coat onto the PM components (e.g., PRRs), which induces concomitant physical changes in the PM endocytic membrane fraction. Once the clathrin coat is loaded onto the cargo, the newly formed compartment gradually bends towards the cytosol, ultimately resulting in a scission from the membrane. While the initial bending force that curves the membrane is provided by the clathrin coated vesicles themselves, the growth and bending of the cargo-containing fraction is driven by actin polymerization. In short, this process is facilitated by the specific attachment of actin to the clathrin coat. Upon binding, the actin filaments extend by polymerizing and branching, a process mediated by the Arp2/3 complex and PM-associated myosin. This process is referred to as actin flow (Kaksonen and Roux, 2018), and it is predicted that plants utilize functionally and mechanistically analogous processes to those in animal systems (Fan et al., 2015).
In the case of plant immune regulation through CME, multiple PRRs, as well as numerous additional PM-associated proteins, have been demonstrated to require CME for plant defense activation and signaling. For example, in the case of PTI, Mbengue et al. (Mbengue et al., 2016) demonstrated that the PRRs FLS2, EFR (Ef-Tu receptor), and PERP1/2 (pep1 receptor) require clathrin, as well as the activity of BRI1-ASSOCIATED KINASE 1 (BAK1), for endocytosis, which are activated by corresponding PAMPs. A second study further indicates that CME is required not only for the endocytosis of PEPR1 itself, but also the activation of PEPR1-mediated defense responses (Ortiz-Morea et al., 2016). Interestingly, myosin inhibitor 2,3-butanedione monoxime (BDM) was found to inhibit FLS2 endocytosis, while the actin filament modifier latrunculin-B (LatB) was shown to have only a minor impact on FLS2 endocytosis (Beck et al., 2012). Taken together, these data support a role for actin cytoskeleton-mediated CME in the turnover and regulation of PM-associated immune receptors and their associated signaling processes. As one might expect, ETI-associated receptors also rely on CME for proper activity, as is the case for the tomato R-protein Cf-4, which functions in immunity against the pathogenic fungus Cladosporium fulvum (Postma et al., 2016).
The role of the cytoskeleton in intercellular trafficking of immune-associated processes
The plant immune response relies on specialized patterns of cellular trafficking to deploy the suite of proteins, organelles, and small molecules required for pathogen resistance signaling (Park et al., 2018). To facilitate the rapid re-localization of immune components to the site of infection, both MF and MT are required for the specific trafficking of immune cargo to the site of infection (Brandizzi and Wasteneys, 2013; Tominaga and Ito, 2015; Nebenfuhr and Dixit, 2018). As a broader function underpinning the regulation of this process, and moreover, the connectivity to PTI, numerous studies have demonstrated that the plant immune signal involves the positive feedback in the expression of PM-cell wall (CW) associated immune components, which include various of signaling complexes, CW-associated polysaccharide synthases, and CW polysaccharide components synthesized in Golgi (Schneider et al., 2016; van de Meene et al., 2017; Bacete et al., 2018). For example, flg22 perception enhances the transcription of FLS2, EFR, BAK1, and RBOHD (Li et al., 2016), a process that is hypothesized to compensate for the turnover (i.e., endocytosis) of PM-associated immune components to sustain the immune (i.e., PTI) signaling capacity of the cell. The enhanced expression these PM-CW localized immune regulators requires a robust cytoskeleton system for their transportation and localization to the membrane. For instance, once pathogen signals (i.e., PAMPs) are perceived, callose-enriched papillae between the CW and PM will form to inhibit pathogen penetration, which is regulated by salicylic and jasmonic acid pathway (Luna et al., 2011; Yi et al., 2014). At a mechanistic level, callose deposition requires callose synthases (CalSs), enzymes that are sorted in Golgi and translocated to the cell wall. This process requires the activity of both MF and MT, and disruption of either cytoskeletal network leads to a dysfunction in CalS (Cai et al., 2011). Accordingly, in another study, it was demonstrated that an Arabidopsis class XI myosin mutant, with disrupted MF/MT trafficking, has dampened callose and lignin accumulation at the fungal infection site (Yang et al., 2014). Thus, from perception of PAMPs to the activation of PTI-associated defense responses, the cytoskeletal network plays a key role in surveillance, activation, and the execution of immunity.
As noted above, the cytoskeleton is also required for the rapid re-localization of various host organelles and proteins to the site of pathogen penetration, a process that is hypothesized to enhance the immune response. In one of the best-characterized examples, Takemoto et al. (Takemoto et al., 2003) observed the accumulation of Arabidopsis ER and Golgi occur at the infection site of oomycete plant pathogen Hyaloperonospora arabidopsidis, simultaneously with rapid remodeling of actin filaments. Subsequent work further showed that these events paralleled the redistribution of the host nucleus, ER, Golgi, mitochondria, and peroxisome at sites adjacent to penetration events during powdery mildew infection (Takemoto et al., 2006; Yang et al., 2014). We posit that these processes function to accelerate defense-associated metabolism, yielding an increase in the rate of response during infection via cytoskeletal-mediated cellular trafficking.
The recent discovery of a role for chloroplast in plant immunity illustrates the complex relationship(s) between immune signaling and the cytoskeletal network. As a component of the plant defense system, the chloroplast plays a role in the activation of HR-PCD though its degradation, which functions as a source of ROS burst following ETI elicitation (Dong and Chen, 2013). Interestingly, disruption of the MT network has been shown to trigger chloroplast autophagy, yet this same disruption attenuates cellular autophagy (Wang et al., 2015). Based on this, it is difficult to discern a role for the concomitant regulation of chloroplast and cytoskeleton as a function of HR-PCD. However, the explanation may lie in recent data describing the function of stromule formation during the activation of plant defense. A recent study found that chloroplasts form a tube-like architecture, called stromules, which stretch towards chloroplasts, other plastids, and even the nucleus, to mediate immune signaling (Hanson and Hines, 2018). As an ETI-associated process, stromules were demonstrated to function in the transport of the N-Receptor Interacting Protein 1 (NRIP1; Caplan et al., 2008) and potentially other pro-immunity molecules into the nucleus to trigger the ETI against tobacco mosaic virus (TMV) effector p50 (Caplan et al., 2015). As a link to the engagement of the cytoskeleton, a recent study confirms that the extension of stromules from the chloroplast is mediated by the cytoskeleton (Kumar et al., 2018). In brief, this work demonstrates that stromules stretch along MT, and application of the MT disrupting agents amiprophos-methyl or oryzalin inhibited the growth of stromules. MF serve as the anchor point rather than the extension track (Kumar et al., 2018), potentially through binding of the stromule via class XI myosin (Natesan et al., 2009). Taken together, these studies provide compelling evidence indicating the deployment of organelles and the transportation of their products is crucial for immune regulation, which relies on the activity of the cytoskeleton.
Battlefield cytoskeleton: the frontline of plant defense and pathogen virulence
Recent data from a suite of studies demonstrate numerous important roles for the plant cytoskeleton in the activation and signaling of plant immunity. However, the question remains: Is the reorganization of the cytoskeleton a response, or a consequence? Is it associated with the activation of immunity, or a process manipulated by pathogens to induce susceptibility? The short answer is both. A leading hypothesis in the field of cell biology and immunity is that the rapid and seemingly random reorganization of the cytoskeletal network is a plant-regulated cellular response to support immune signaling and downstream signaling of defense (Day et al., 2011). In this case, recent data demonstrates that pathogens alter both types of cytoskeletal structures during infection to evade immunity and promote infection. In the case of the former, recent work has shown that rapid changes in cytoskeletal organization occur during immune activation. For example, perception of the PAMPs flg22, elf26, and chitin have all been shown to trigger the reorganization of actin in Arabidopsis epidermal cells (Henty-Ridilla et al., 2013; Henty-Ridilla et al., 2014) and in stomatal guard cells (Shimono et al., 2016a). As predicted, these PAMP-stimulated events require the PRRs FLS2, Ef-Tu, and CERK1, reinforcing the requirement of the actin cytoskeleton for PRR-PAMP function. Upon infection of tobacco BY-2 cells, the Pseudomonas syringae DC3000 (Pst DC3000) type III secretion system (T3SS) helper protein, HrpZ, has been demonstrated to function as a PAMP, the perception of which induces bundling of F-actin and a concomitant decrease in MT density (Guan et al., 2013).
Alternatively, it is also demonstrated that pathogens can alter actin cytoskeletal structures during infection to evade immunity and promote infection. In a follow-up infection assay using Pst DC3000, it was observed that while the MT architecture did not change within 16 hpi (Lee et al., 2012b); treatment for longer periods (i.e., >20 h) tended to induce long-term and multiplephase influences on host actin. These changes included an initial increase in MF density, followed by a decrease in MF density with a concomitant increase in MF bundling at later stages of infection (Henty-Ridilla et al., 2013). Importantly, a type-III secretion system (T3SS)-deficient, avirulent, strain Pst DC3000 ∆hrpH was unable to trigger the second phase of remodeling, suggesting a role of pathogen virulence by the T3SS as well as T3Es themselves (Shimono et al., 2016b).
In the case of fungal pathogens, similar to bacteria, avirulent and virulent strains confer differences in the pattern of cytoskeleton re-organization, illustrating a role for the cytoskeleton as a common immune component in response to multiple types of pathogens. In the well-defined barley-powdery mildew interaction system, avirulent strains will trigger the rapid reorganization of host MF and MT during the invasion process (Kobayashi et al., 1992; Opalski et al., 2005; Miklis et al., 2007); this response is indicated by actin bundling at the interface of the mature appressorium, with the formation of a dense network of MF surrounding the papillae. Such phenomena are referred to as actin focusing, with F-actin linking the host nucleus and the host-appressorium interface. For virulent strains, however, this pattern of filament organization is not observed, with only a slight aggregation of filament bundles without actin focusing (Kobayashi et al., 1992; Opalski et al., 2005; Miklis et al., 2007). Interestingly, MT remodeling patterns show a similar trend, with the induction of thick radial arrays of MT bundles at the site of appressorium formation in the presence of avirulent isolates and no aggregation in the presence of virulent strains (Kobayashi et al., 1992). Similar to powdery mildew, studies in the cowpea-rust fungi interaction system also demonstrated that avirulent strains trigger MF and MT reorganization, leading to a reduction in filament density, while no significant reorganization is observed in cells infected by virulent strains (Skalamera and Heath, 1998).
In the case of bacterial pathogen infection, this phenomenon can be phenocopied by the application of cytoskeletal agents that interfere with MF and MT dynamics, manifesting in differing immune phenotypes between bacterial and fungal pathogens. For example, in the case of bacterial phytopathogens, disrupted MF increases resistance, including both PTI and ETI branches (Tian et al., 2009; Henty-Ridilla et al., 2013; Kang et al., 2014; Krutinova et al., 2018), while disrupted MT increase susceptibility to infection (Lee et al., 2012b). However, host resistance to fungal pathogens is usually dampened by both MF and MT dynamics inhibitor (Schmidt and Panstruga, 2007). These data indicate that cytoskeletal architecture has a significant influence on plant immunity, potentially controlled by both host and pathogen to alter the balance of resistance versus susceptibility.
While the broader function and mechanism(s) associated with MF/MT (re)organization in response to pathogen infection remain largely undefined, insight into the role of the cytoskeleton in plant immunity is becoming clearer through the analysis of individual MF- and MT-associated proteins. Among the first regulators of actin cytoskeletal organization revealed to play an important role in immunity are the actin depolymerizing factor (ADF)/cofilin (hereafter referred to as AC) family of proteins – a conserved class of small proteins that regulate actin cytoskeletal organization via filament severing and depolymerization (Kanellos and Frame, 2016). As a family, ACs are widely conserved across all eukaryotes, yet their abundance varies: In mammals, 3 ACs have been identified (i.e., ADF, CFL1, and CFL2) and in most plants, dozens of ADF-encoding genes are present (11 in Arabidopsis, up to 27 in banana) (Kanellos and Frame, 2016; Nan et al., 2017). Similar to their mammalian counterparts, plant ADFs function as key regulators of cytoskeletal organization, controlling the overall balance of cellular G- and F-actin ratios.
In recent studies, ADFs have also been shown to be associated with the function and activity of the plant immune system. For example, as a regulator of PTI, it was demonstrated that Arabidopsis ADF4 plays a key role in PAMP-triggered actin remodeling, demonstrating that ADF4 – and actin depolymerization – are necessary components of actin remodeling and callose deposition upon elf26 perception by the EFR (Henty-Ridilla et al., 2014). Moreover, in the case of fungal pathogen perception and immunity, the adf4 mutant was found to possess enhanced resistance, with subclass I ADFs imparting an additive effect on pathogen susceptibility (Inada et al., 2016). These data suggest that resistance signaling associated with ADF function may in fact be mediated in a homologue/class-specific manner, and moreover, that expansion of the ADF gene family in plants, as compared to mammals, may impart roles for specific and individual ADFs. Indeed, additional data support this hypothesis: Arabidopsis ADF6 was shown to negatively regulate the localization of RPW8.2 to extrahaustorial membranes to promote immune signaling (Wang et al., 2009); ADF3 is a positive regulator of resistance against aphids (Mondal et al., 2017); and in wheat, TaADF4 and TaADF7 significantly contribute to resistance against the stripe rust pathogen Puccinia striiformis (Fu et al., 2014; Zhang et al., 2017), while TaADF3 is an negative regulator of this interaction (Tang et al., 2015). In addition to ADFs, the roles of other MF/MT associated proteins in plant immunity are beginning to emerge. For instance, a recent study demonstrated that Arabidopsis Profilin3 (PFN3) negatively regulates PTI by inhibiting formin-mediated actin polymerization (Sun et al., 2018).
Pathogen targeting of cytoskeletal organization: immune subversion and pathogenicity
Given the incredible connectivity of the cytoskeletal platform to nearly all cellular networks (Figure 2), it is not surprising that pathogens and pests have evolved mechanisms to block immunity – either directly or indirectly – through manipulation of cytoskeletal function. In this respect, by targeting a few key steps in cytoskeletal assembly, for example, pathogens can gain access to a range of host mechanisms. To usurp, evade, or destroy? These are the evolved “choices” that pathogens have made to overtake the function and activity of the immune system at the cytoskeletal interface. In the case of plant viruses, whose amplification and intercellular movement require manipulation of the host cell machinery, including cytoskeleton (Hong and Ju, 2017), the “choice” is to usurp. As a general strategy for viral manipulation of the cytoskeleton, the viral replication complex (VRC) can load itself onto the cytoskeleton using scaffold proteins (e.g., movement protein, linking protein) or myosins, which enable the virus to track along the cytoskeletal network, including through plasmodesmata (Pitzalis and Heinlein, 2017). As a result, the infecting virus is able to move from cell to cell, overwhelming immunity, and ultimately taking control of the host.
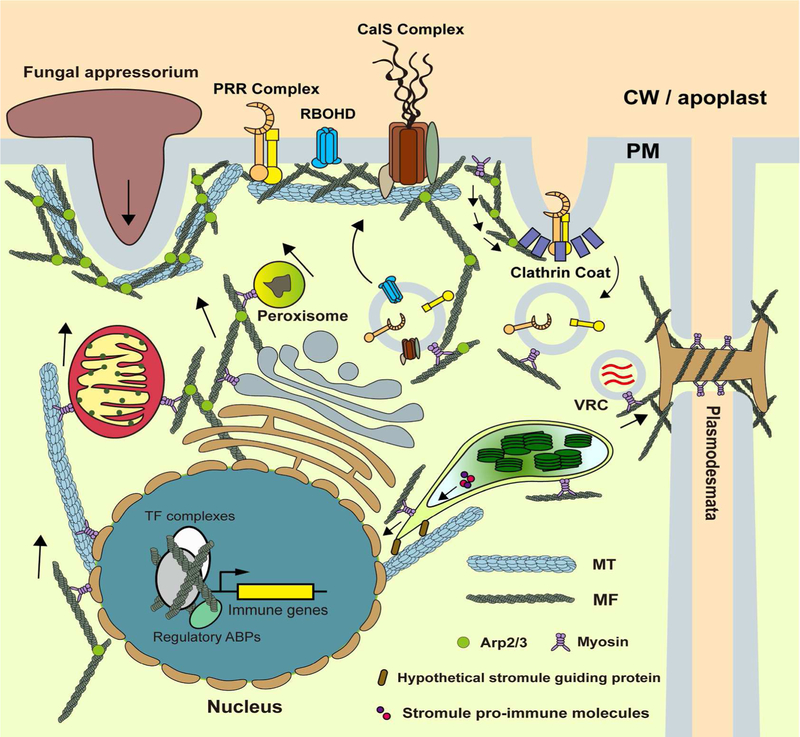
Fig. 2.
The versatility and involvement of the plant cytoskeleton in immunity. Plant MF and MT are involved in multiple processes during the immune response. The cytoskeleton provides the physical attachment, as well as specialized microenvironments, to numerous PM-associated immune processes (e.g., PRR complexes, RBOHD, and CalS complex) and is required for full functionality of these immune processes. The cytoskeleton is also required pro-immune cellular trafficking, a process that is associated with the transport of organelles, proteins, and small molecules, through the action of endocytosis, plastid stromules, as well as cell-to-cell trafficking through plasmodesmata. Virus replication complex (VRC) can hijack cytoskeleton and transports to adjacent cells through plasmodesmata. As a less-characterized mechanism of plant immunity, actin is also involved in the transcriptional regulation of immune signaling events within the nucleus, potentially through aiding in the formation of a regulatory complex consisting of transcription factors and chromatin. Arrows are shown to indicate the movement of corresponding cellular components.
As noted above, pathogen effector molecules function to subvert immune signaling, and in recent years, much effort has been spent on the discovery of the constellation of host processes targeted by these secreted factors. Thus, it was only a matter of time before pathogen effectors were identified which can directly and/or indirectly influence cytoskeletal function. In the case of indirect modulation of cytoskeletal function, work from Lee and colleagues (Lee et al., 2012b) observed that the Pst DC3000 T3E HopE1 can bind to calmodulin, a process that leads to disassociation of the microtubule-associated protein 65–1 (MAP65–1) from the MT network, resulting in an increase in susceptibility to Pst DC3000. In a similar mechanism, the Xanthomonas euvesicatoria T3E AvrBsT, and acetyltransferase, was shown to acetylate ACETYLATED INTERACTING PROTEIN1 (ACIP1), causing it to dissociate from MT, leading to a dampening of plant immunity (Cheong et al., 2014). Interestingly, it has also been demonstrated that pathogenic effectors can also influence the regulation of cytoskeletal function within and between organelles, as demonstrated by Erickson et al., which demonstrated that the Xanthomonas campestris T3E XopL suppresses plastid stromule formation during immune signaling by targeting unknown MT-associated proteins (Erickson et al., 2018).
Lastly, and in work supported by independent studies that converged on similar pathogen virulence mechanisms, is the case of the P. syringae T3E HopG1. Previous work showed that HopG1 is a mitochondria-targeted effector that suppresses plant immunity (Block et al., 2010). In a bid to define virulence factors that target host cytoskeletal immune signaling, Shimono et al. (Shimono et al., 2016b) demonstrated that HopG1 interacts with Arabidopsis kinesin 7.4 (i.e., Kin7.4; Moschou et al., 2016), a mitochondria-localized motor protein (Itoh et al., 2001) whose function is required for actin filament organization. During Pst DC3000 infection of Arabidopsis, HopG1 is delivered into the host cell, and subsequently associates with Kin7.4, resulting in actin bundling and enhanced disease symptom development. This is exciting from the standpoint of pathogen targeting of the cytoskeleton and in the broader context of a role for actin in immunity. For example, if HopG1 and kinesin associate on the mitochondrial outer membrane, this might suggest a mechanism to inhibit the motor activity of kinesin, leading to an impediment in mitochondrial motion through the concerted action of both (i.e., MF and MT) cytoskeletal networks. This would then lead to a reduction in the energy needed to support cytoskeletal function and dynamism (Bartolák-Suki et al., 2017). However, if HopG1 and kinesin localize within the mitochondria itself, it would indicate a role for HopG1 in the disruption of kinesin function and a broader role of mitochondria as a signaling hub for immunity and cell death through actin filament remodeling. We tend to favor the latter, as evidence for such a role is supported by numerous studies demonstrating a function for the actin-mitochondrial network as a hub for the activation of apoptosis (reviewed in Elmore, 2007), a process associated with pathogen-induced senescence (Shimono et al., 2016b).
Converse to the examples highlighted above – whereby pathogens finesse cytoskeletal function to promote infection and disease – pathogens have also evolved virulence strategies to disrupt actin cytoskeletal dynamics in a more abrupt manner. In short, pathogens can paralyze host immunity by directly dissembling the cytoskeletal machinery. In one of the first examples of directed targeting of the plant cytoskeleton by a phytopathogen, work by Lee et al. (Lee et al., 2012a) uncovered a mechanism whereby the P. syringae T3E HopZ1a, an acetyltransferase, can modify and disrupt the MT network to interfere with MT-supported processes, such as trafficking. More recently, work by Kang et al. (Kang et al., 2014) showed that the P. syringae T3E HopW1 disrupts F-actin integrity by directly depolymerizing actin filaments during infection, a process resulting in blocks to protein cargo trafficking and endocytosis. Such strategies are also employed by viral pathogens, such as the case of the cucumber mosaic virus (CMV) movement protein (MP), which can sever F-actin to increase the size exclusion limit of plasmodesmata, potentially accelerating the viral spread to adjacent cells (Su et al., 2010). Additionally, a newest study demonstrates that root-knot nematodes secrets an effector, Meloidogyne incognita Profilin3 (MiPFN3) into host “giant cell”, the feeding structure, to inhibit host actin polymerization and cause higher susceptibility (Leelarasamee et al., 2018).
While each of these examples clearly demonstrates that direct disruption of the MT or MF networks are strategies to impede plant immunity, the question remains as to how the activities of these effectors are coordinately regulated, since absolute disruption of plant MF/MT does not always lead to attenuated immune response, as mentioned above. One hypothesis is that additional signals are generated during infection that lead to proper regulation of effector-mediated cytoskeleton dissemble. This would, hypothetically, result in the specific modulation of effectors’ activities at different key stages of the infection process, which leads to disruption of immune signaling. Such strategies have been characterized in the case of Salmonella infection of human cells, in which the co-regulation of the type-III effectors SipA and SipC, with opposing yet cooperative, actin polymerizing and depolymerizing activites renders the immune escape (McGhie et al., 2001; McGhie et al., 2009).
Emerging themes and future directions: The PTI, ETI, actin frontier
The past two decades have witnessed the discovery of numerous mechanisms underpinning the linkages between the cytoskeleton and plant immune system (Figure 3). While a number of mechanisms remain to be defined, a framework is starting to emerge that demonstrates the role(s) of MF and MT in processes associated with PTI, ETI, and pathogen virulence. As a roadmap for future research in this area, we believe that following topics will be key in further defining the actin-pathogen connection:
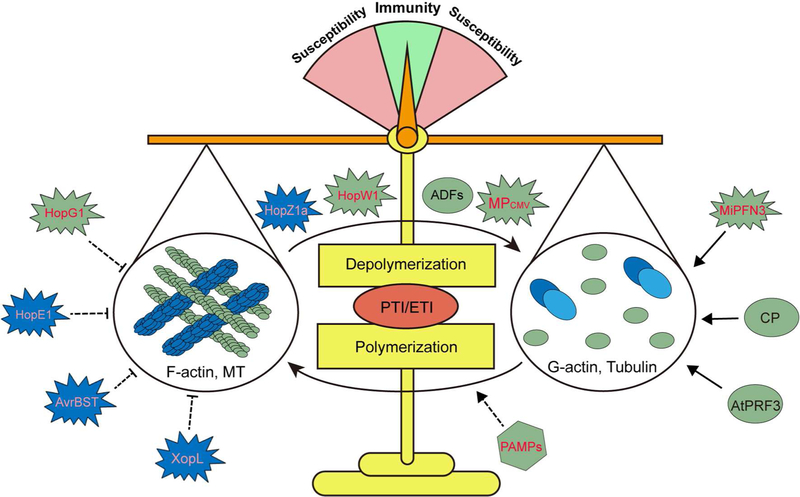
Fig. 3.
The cytoskeleton is central to the balance between immunity and susceptibility, and the host-dominated balance and alterability between G-actin, F-actin, and tubulin control the plant response to pests and pathogens. The cytoskeleton is tightly regulated by the temporal and spatial control of filament architecture, and these points of control are influenced by the perception of pathogens and pathogen elicitors (i.e., PAMPs, effectors). For host, identified actin regulator in immune response includes ADFs (depolymerizing and severing actin), CP (stabilizing short F-actin oligo and G-actin), and AtPFR3 (stabilizing G-actin). Pathogen and pests, on the other hand, can use effectors to interfere the host regulation of cytoskeleton. For instance, HopW1 and the CMV movement protein (MP) can directly sever F-actin, thus increasing the cellular concentration of G-actin. Similarly, HopZ1a can disrupt MT filaments by acetylating tubulins, a process that results in disruption of the MT network and associated process. In the case of MF function, MiPFN3 can stabilize G-actin and directly inhibit actin assembly. HopG1, HopE1, AvrBsT, and XopL can indirectly interfere host cytoskeletal function.
Pathogen perception. How is plant immunity regulated by PM-associated cytoskeleton? While PRR (e.g., FLS2) activation is not inhibited by disrupting F-actin, the ROS burst intensity and response time are altered. Given that RBOHD activation by PRR-complexes does require a function association with the cytoskeleton, the most obvious hypothesis is that PM-associated actin potentially functions as a scaffold for PRR regulatory complexes. Such mechanisms in plants, if they exist, may parallel what is currently described in innate immune signaling processes in animals.
PTI versus ETI. At what points of convergence between PTI and ETI does actin function? At present, supporting data for a role of actin in both PTI and ETI exists, but does not provide much detail as to their function and regulation. Additionally, it will be important to determine if PM-associated cytoskeleton influences ETI through CC-NLR ETI activation. The convergence of actin on immune receptor function will be an important discovery in defining the immune-membrane function.
Regulation of actin, and pathogen targeting of the cytoskeleton. What is the signaling pathway from PRR activation to resultant cytoskeleton reorganization? An abundance of data supports a critical role for ADF in immunity to plant pathogens. However, a gap remains as to how ADF activity is regulated in plants. In animal systems, cofilin regulation is intimately linked to the function of the immune system; it is predictable that such mechanism is also adapted by plant. At the same time, it is possible that pathogens can target the ADF-actin switch, which leads to misregulation of the cytoskeleton and further blocks immune signaling and host defense.
From the outside, in. What is the function of actin system inside the nucleus? The extraordinary connectivity of the cytoskeleton gives the cell unfettered access to a range of processes and environments. Further definition of how signals are transduced from the PM to the nucleus will provide insight into the surveillance and regulatory functions of the immune system as a continuum, from the periphery of the cell to the nucleus. In mammal systems, this process is well documented (Wada et al., 1998; Stuven et al., 2003; Dopie et al., 2012). Similar to the operation of the plant immune system, the nuclear-cytoplasmic shuttling of actin is hypothesized to maintain an active, and highly responsive surveillance platform. In plants, preliminary studies have uncovered a role for nuclear-cytoplasmic shuttling of actin during pathogenesis (Levy et al., 2013), and nuclear ABPs, such as ADFs (Inada et al., 2016), have an impact on the plant immune response. Thus, the framework exists to further define the processes described here towards extending our understanding of the role of the cytoskeleton in almost every step of plant immune response, from pathogen perception to the regulation of the immune transcriptome.
Acknowledgements
We thank the members of the Day laboratory and Dr. Noel Day for useful comments and feedback during the preparation of this manuscript. Research from the Day lab summarized herein was supported by grants from the U.S. National Science Foundation (IOS-1557437) and the National Institutes of Health (1R01GM125743) to B.D.
References Cited
- Bacete L, Melida H, Miedes E, and Molina A 2018. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J 93:614–636. [DOI] [PubMed] [Google Scholar]
- Bartolák-Suki E, Imsirovic J, Nishibori Y, Krishnan R, and Suki B 2017. Regulation of mitochondrial structure and dynamics by the cytoskeleton and mechanical factors. Int J Mol Sci 18:1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Zhou J, Faulkner C, MacLean D, and Robatzek S 2012. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24:4205–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielig H, Lautz K, Braun PR, Menning M, Machuy N, Brugmann C, Barisic S, Eisler SA, Andree M, Zurek B, Kashkar H, Sansonetti PJ, Hausser A, Meyer TF, and Kufer TA 2014. The cofilin phosphatase slingshot homolog 1 (SSH1) links NOD1 signaling to actin remodeling. PLoS Pathog 10:e1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Guo M, Li G, Elowsky C, Clemente TE, and Alfano JR 2010. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol 12:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, and Wasteneys GO 2013. Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J 75:339–349. [DOI] [PubMed] [Google Scholar]
- Bucherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, and Zipfel C 2017. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6: e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D 2016. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol Rev 40:894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Faleri C, Del Casino C, Emons AM, and Cresti M 2011. Distribution of callose synthase, cellulose synthase, and sucrose synthase in tobacco pollen tube is controlled in dissimilar ways by actin filaments and microtubules. Plant Physiol 155:1169–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, and Dinesh-Kumar SP 2008. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Kumar AS, Park E, Padmanabhan MS, Hoban K, Modla S, Czymmek K, and Dinesh-Kumar SP 2015. Chloroplast stromules function during innate immunity. Dev Cell 34:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Kirik A, Kim JG, Frame K, Kirik V, and Mudgett MB 2014. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog 10:e1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, and Staskawicz BJ 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814. [DOI] [PubMed] [Google Scholar]
- Couto D, and Zipfel C 2017. Regulation of pattern recognition receptor signaling in plants. Nat Rev Immunol 16:537–552. [DOI] [PubMed] [Google Scholar]
- Day B, Henty JL, Porter KJ, and Staiger CJ 2011. The pathogen-actin connection: A platform for defense signaling in plants. Annu Rev Phytopathol 49:483–506. [DOI] [PubMed] [Google Scholar]
- Dong J, and Chen W 2013. The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. PLoS One 8:e73091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Skarp K-P, Rajakyla EK, Tanhuanpaa K, and Vartiainen MK 2012. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A 109:3205–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott A, and Shaw SL 2018. Update: Plant cortical microtubule arrays. Plant Physiol 176:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S 2007. Apoptosis: A review of programmed cell death. Toxicol Pathol 35:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JL, Adlung N, Lampe C, Bonas U, and Schattat MH 2018. The Xanthomonas effector XopL uncovers the role of microtubules in stromule extension and dynamics in Nicotiana benthamiana. Plant J 93:856–870. [DOI] [PubMed] [Google Scholar]
- Fan L, Li R, Pan J, Ding Z, and Lin J 2015. Endocytosis and its regulation in plants. Trends Plant Sci 20:388–397. [DOI] [PubMed] [Google Scholar]
- Fu Y, Duan X, Tang C, Li X, Voegele RT, Wang X, Wei G, and Kang Z 2014. TaADF7, an actin‐depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J 78:16–30. [DOI] [PubMed] [Google Scholar]
- Guan X, Buchholz G, and Nick P 2013. The cytoskeleton is disrupted by the bacterial effector HrpZ, but not by the bacterial PAMP flg22, in tobacco BY-2 cells. J Exp Bot 64:1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, and Hines KM 2018. Stromules: Probing formation and function. Plant Physiol 176:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Huang T, Ao K, Yan X, and Huang Y 2017. Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by ubiquitination. Front Plant Sci 8:1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Li JJ, Day B, and Staiger CJ 2014. ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 26:340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Shimono M, Li JJ, Chang JH, Day B, and Staiger CJ 2013. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog 9: e1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-S, and Ju H-J 2017. The plant cellular systems for plant virus movement. Plant Pathol J 33:213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, and He SY 2014. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7:1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Lema AS, Coll NS, and Nowack MK 2017. Dying two deaths - programmed cell death regulation in development and disease. Curr Opin Plant Biol 35:37–44. [DOI] [PubMed] [Google Scholar]
- Inada N, Higaki T, and Hasezawa S 2016. Nuclear function of subclass I actin depolymerizing factor contributes to susceptibility in Arabidopsis to an adapted powdery mildew fungus. Plant Physiol 170:1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, and Yoshida S 2001. Kinesin-related proteins with a mitochondiral targeting signal. Plant Physiol 127:724–726. [PMC free article] [PubMed] [Google Scholar]
- Jones JD, and Dangl JL 2006. The plant immune system. Nature 444:323–329. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, and Roux A 2018. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19:313–326. [DOI] [PubMed] [Google Scholar]
- Kanellos G, and Frame MC 2016. Cellular functions of the ADF/cofilin family at a glance. J Cell Sci 129:3211–3218. [DOI] [PubMed] [Google Scholar]
- Kang Y, Jelenska J, Cecchini NM, Li Y, Lee MW, Kovar DR, and Greenberg JT 2014. HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog 10:e1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, and Panstruga R 2010. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285:39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I, Kobayashi Y, Yamaoka N, and Kunoh H 1992. Recognition of a pathogen and a nonpathogen by barley coleoptile cells. III. Responses of microtubules and actin filaments in barley coleoptile cells to penetration attempts. Can J Bot 70:1815–1823. [Google Scholar]
- Krutinova H, Trda L, Kalachova T, Lamparova L, Pospichalova R, Dobrev P, Malinska K, Burketova L, Valentova O, and Janda M 2018. Can actin depolymerization actually result In increased plant resistance to pathogens? bioRxiv:278986. [DOI] [PMC free article] [PubMed]
- Kufer TA, Kremmer E, Adam AC, Philpott DJ, and Sansonetti PJ 2008. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol 10:477–486. [DOI] [PubMed] [Google Scholar]
- Kumar AS, Park E, Nedo A, Alqarni A, Ren L, Hoban K, Modla S, McDonald JH, Kambhamettu C, Dinesh-Kumar SP, and Caplan JL 2018. Stromule extension along microtubules coordinated with actin-mediated anchoring guides perinuclear chloroplast movement during innate immunity. Elife 7:e23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YC, Bornke F, Angers S, Wilde A, Guttman DS, and Desveaux D 2012a. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog 8:e1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AHY, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YLC, Bornke F, Angers S, Wilde A, Guttman DS, and Desveaux D 2012b. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog 8:e1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelarasamee N, Zhang L, and Gleason C 2018. The root-knot nematode effector MiPFN3 disrupts plant actin filaments and promotes parasitism. PLoS Pathog 14:e1006947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand-Poels S, Kustermans G, Bex F, Kremmer E, Kufer TA, and Piette J 2007. Modulation of Nod2-dependent NF-kappaB signaling by the actin cytoskeleton. J Cell Sci 120:1299–1310. [DOI] [PubMed] [Google Scholar]
- Levy A, Zheng JY, and Lazarowitz SG 2013. The tobamovirus turnip vein clearing virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J Virol 87:6428–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, and He P 2016. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, and Kahmann R 2017. How filamentous plant pathogen effectors are translocated to host cells. Curr Opin Plant Biol 38:19–24. [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, and Ton J 2011. Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24:183–193. [DOI] [PubMed] [Google Scholar]
- Mbengue M, Bourdais G, Gervasi F, Beck M, Zhou J, Spallek T, Bartels S, Boller T, Ueda T, Kuhn H, and Robatzek S 2016. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci U S A 113:11034–11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, and Koronakis V 2001. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J 20:2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, and Koronakis V 2009. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, and Panstruga R 2007. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144:1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal HA, Louis J, Archer L, Patel M, Nalam VJ, Sarowar S, Sivapalan V, Root DD, and Shah J 2017. Arabidopsis ACTIN-DEPOLYMERIZING FACTOR3 is required for controlling aphid feeding from the phloem. Plant Physiol 176: 879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou PN, Gutierrez-Beltran E, Bozhkov PV, and Smertenko A 2016. Separase promotes microtubule polymerization by activating CENP-E-related kinesin Kin7. Dev Cell 37:350–361. [DOI] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, and Yang Z 2012. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10:e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Q, Qian D, Niu Y, He Y, Tong S, Niu Z, Ma J, Yang Y, An L, and Wan D 2017. Plant actin-depolymerizing factors possess opposing biochemical properties arising from key amino acid changes throughout evolution Plant Cell 29:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, and Gray JC 2009. Myosin XI is required for actin-associated movement of plastid stromules. Mol Plant 2:1262–1272. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A, and Dixit R 2018. Kinesins and myosins: molecular motors that coordinate cellular functions in plants. Annu Rev Plant Biol 69:329–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, and Hückelhoven R 2005. The receptor‐like MLO protein and the RAC/ROP family G‐protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f. sp. hordei. Plant J 41:291–303. [DOI] [PubMed] [Google Scholar]
- Ortiz-Morea FA, Savatin DV, Dejonghe W, Kumar R, Luo Y, Adamowski M, Van den Begin J, Dressano K, Pereira de Oliveira G, Zhao X, Lu Q, Madder A, Friml J, Scherer de Moura D, Russinova E 2016. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci U S A 113:11028–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Nedo A, Caplan JL, and Dinesh-Kumar SP 2018. Plant-microbe interactions: organelles and the cytoskeleton in action. New Phytol 217:1012–1028. [DOI] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, and Zhang Y 2018. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact 31:403409. [DOI] [PubMed] [Google Scholar]
- Pitzalis N, and Heinlein M 2017. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J Exp Bot 69:117–132. [DOI] [PubMed] [Google Scholar]
- Porter K, and Day B 2015. From filaments to function: The role of the plant actin cytoskeleton in pathogen perception, signaling, and immunity. J Integ Plant Biol 58:299–311. [DOI] [PubMed] [Google Scholar]
- Porter KJ, Shimono M, Tian M, and Day B 2012. Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog 8:e1003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Liebrand TW, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MH, and Robatzek S 2016. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 210:627–642. [DOI] [PubMed] [Google Scholar]
- Schmidt SM, and Panstruga R 2007. Cytoskeleton functions in plant–microbe interactions. Physiol Mol Plant Path 71:135–148. [Google Scholar]
- Schneider R, Hanak T, Persson S, and Voigt CA 2016. Cellulose and callose synthesis and organization in focus, what’s new? Curr Opin Plant Biol 34:9–16. [DOI] [PubMed] [Google Scholar]
- Shimono M, Higaki T, Kaku H, Shibuya N, Hasezawa S, and Day B 2016a. Quantitative evaluation of stomatal cytoskeletal patterns during the activation of immune signaling in Arabidopsis thaliana. PLoS One 11:e0159291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Lu YJ, Porter K, Kvitko BH, Henty-Ridilla JL, Creason A, He SY, Chang JH, Staiger CJ, and Day B 2016b. The Pseudomonas syringae type-III effector HopG1 induces actin remodeling to promote symptom development and susceptibility during infection. Plant Physiol 171: 2239–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalamera D, and Heath MC 1998. Changes in the cytoskeleton accompanying infectioninduced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. Plant J 16:191–200. [DOI] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, and Gorlich D 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin-actin complexes. EMBO J 22:5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Liu Z, Chen C, Zhang Y, Wang X, Zhu L, Miao L, Wang X-C, and Yuan M 2010. Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22:1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Spears BJ, Kim SH, and Gassmann W 2018. Constant vigilance: plant functions guarded by resistance proteins. Plant J 93:637–650. [DOI] [PubMed] [Google Scholar]
- Sun H, Qiao Z, Chua KP, Tursic A, Liu X, Gao YG, Mu Y, Hou X, and Miao Y 2018. Profilin negatively regulates formin-mediated actin assembly to modulate PAMPtriggered plant immunity. Curr Biol 28:1882–1895. [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, and Hardham AR 2003. GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33:775–792. [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, and Hardham AR 2006. Re-organization of the cytoskeleton and endoplasmic reticulum in the Arabidopsis pen1–1 mutant inoculated with the nonadapted powdery mildew pathogen, Blumeria graminis f. sp. hordei. Mol Plant Pathol 7:553–563. [DOI] [PubMed] [Google Scholar]
- Tang C, Deng L, Chang D, Chen S, Wang X, and Kang Z 2015. TaADF3, an actindepolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front Plant Sci 6:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, and Zhou JM 2017. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29:618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, and Day B 2009. Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol 150:815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, and Ito K 2015. The molecular mechanism and physiological role of cytoplasmic streaming. Curr Opin Plant Biol 27:104–110. [DOI] [PubMed] [Google Scholar]
- Toth K, and Stacey G 2015. Does plant immunity play a critical role during initiation of the legume-rhizobium symbiosis? Front Plant Sci 6:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Meene AM, Doblin MS, and Bacic A 2017. The plant secretory pathway seen through the lens of the cell wall. Protoplasma 254:75–94. [DOI] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, and Nishida E 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J 17:1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WM, Wen YQ, Berkey R, and Xiao SY 2009. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21:2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zheng X, Yu B, Han S, Guo J, Tang H, Yu AY, Deng H, Hong Y, and Liu Y 2015. Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy 11:2259–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Qin L, Liu G, Peremyslov VV, Dolja VV, and Wei Y 2014. Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc Natl Acad Sci U S A 111:13996–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SY, Shirasu K, Moon JS, Lee SG, and Kwon SY 2014. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS One 9:e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Hua Y, Wang J, Huo Y, Shimono M, Day B, and Ma Q 2017. TaADF4, an actin‐depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. Plant J 89:1210–1224. [DOI] [PubMed] [Google Scholar]