Abstract
Cervicovaginal bacteria cause inflammation which in turn increases HIV risk. Profiling the cervicovaginal microbiome, therefore, is instrumental for vaccine development. We show that the microbiome profile captured by cervicovaginal lavage is comparable to samples obtained by vaginal swabs. Thus, lavage may serve as a sampling strategy in NHP vaccine studies.
Keywords: 16S rRNA, HIV, swab, lavage
INTRODUCTION
The microbiology of the lower female genital tract (FGT) influences HIV transmission risk in women and may impact HIV vaccine efficacy(1–3). Therefore, FGT microbiome analysis is necessary in pre-clinical HIV vaccine studies, particularly in vaginal challenge models, to rigorously interpret vaccine efficacy.
An impediment to routine FGT microbiome profiling is the difficulty in obtaining a sufficient amount of vaginal secretion via swabbing to permit accurate assessment of both the mucosal microbiome and vaccine-elicited antibody titers, particularly in older animals. Cervicovaginal lavage (CVL) would therefore be the preferred technique. While a few studies in rhesus have employed CVL to analyze FGT microbial composition, determining whether bacterial communities captured by CVL are sufficiently similar to that captured by swabs is important to support routine incorporation of CVL for more efficient microbiome profiling(4, 5).
Our data demonstrate that mucosa-associated microbiome profiling from CVL is representative of that obtained by swab allowing for swabs to be prioritized for humoral assays and CVL for microbiome analysis. These findings support the adoption of CVL for efficient FGT microbiome sampling in NHP vaccine studies.
MATERIALS AND METHODS
Animals and sampling
Four adult menstruating females were selected from the indoor rhesus colony at the California National Primate Research Center (CNPRC) (Figure 1A, B). Selected animals did not receive oral antibiotic treatment or drugs for at least 4 weeks prior to sampling. Animals were sampled under anesthesia for collection of paired vaginal swabs and cervicovaginal lavages. Briefly, vaginal swabs were collected by Weck-Cel eye spear sponges premoistened with 50μL of saline and inserted for 5 minutes to collect secretions. Subsequently, the perineal area was thoroughly cleaned with saline soaked gauze pads. One ml of PBS was instilled into the vagina using a sterile 1 cc syringe. The fluid was then aspirated and re-instilled gently several times. Swabs were held on dry ice and CVL on ice until processing which occurred within three hours of collection. Briefly, CVL samples were centrifuged at 1500 rpm for 5 min at 4°C to separate cellular fraction from the supernatant as described by us previously(6). The cell pellet and supernatant were separated and immediately flash frozen on dry ice and stored at −80°C until DNA extraction. The Institutional Animal Care and Use Committee (IACUC) of the CNRPC (assurance # 18747) reviewed and approved all animal studies.
Figure 1.
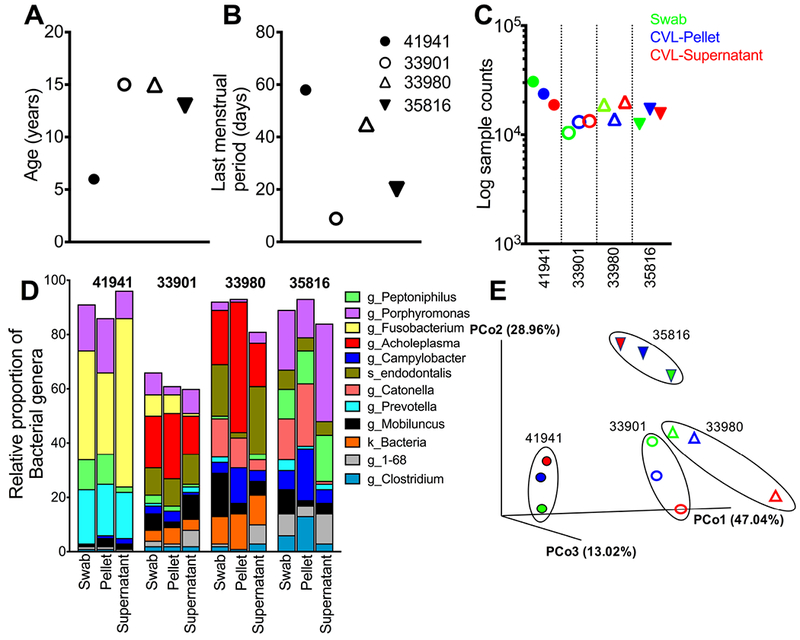
Comparison of sampling methods for profiling rhesus vaginal microbiome. (A) Age of animals sampled, (B) days since last menstrual period and (C) sample counts identified by 16S rRNA V3-V4 sequencing of vaginal swabs (green), CVL supernatant (blue), and CVL pellet (red). (D) Relative abundance of bacterial genera and (E) shows Principal Coordinate analysis using the weighted UniFrac distance metric.
Sequencing of vaginal microbial communities
DNA was isolated using the Qiagen DNeasy PowerSoil kit (Qiagen, Germantown, MD) and primer pairs 319F/806R were used to amplify the V3-V4 domains of the 16S rRNA using a two-step PCR procedure. In step one, both forward and reverse primers contained an Illumina tag sequence, a variable length spacer, a linker sequence, and the 16S target sequence to increase diversity and improve the quality of the sequencing and run. Each 25 μl PCR reaction contained 1 unit Kapa2G Robust Hot Start Polymerase (Kapa Biosystems, Wilmington, MA), 1.5 mM MgCl2, 0.2 mM dNTP mix, 0.2 μM of each primer and 1μl of DNA for each sample.
In step two, each sample was barcoded with a unique forward and reverse barcode combination with an Illumina P5 adapter sequence, a unique 8 nt barcode, a partial matching sequence of the forward adapter used in step one, and reverse primers with an Illumina P7 adapter sequence, unique 8 nt barcode, and a partial matching sequence of the reverse adapter used in step one. The PCR reaction in step two contained 0.2 μM final concentration of each uniquely barcoded primer and 1μl of the product from the PCR reaction in step one.
The final product was quantified on the Qubit instrument using the Qubit Broad Range DNA kit (Invitrogen, Carlsbad, CA) and individual amplicons were pooled in equal concentrations. The pooled library was cleaned utilizing Ampure XP beads (Beckman Coulter, Brea, CA), and the band of interest was subjected to isolation via gel electrophoresis on a 1.5% gel (Sage Science, Beverly, MA). The library was quantified via qPCR followed by 300-bp paired-end sequencing using an Illumina MiSeq instrument in the Genome Center DNA Technologies Core at UC Davis.
Taxonomic classification and bioinformatics analysis
Demultiplexing of the Raw FASTQ files and adapter trimming of sequences were performed using dbcAmplicons version 0.8.5 (https://github.com/msettles/dbcAmplicons). The unmerged forward and reverse reads were imported into QIIME2 version 2017.12 (https://qiime2.org) and sequence variants were determined following the DADA2 analysis pipeline. Measures of β-diversity were generated with weighted UniFrac analysis and the resulting distance matrix was used to perform Principal Coordinate Analysis (PCoA)(7). Taxonomic classification was assigned using a Naive Bayes filtered classifier trained on the 99% identity Green genes database, version 13_8.
RESULTS
The purpose of this study was to determine whether representation of bacterial communities captured by CVL was comparable to that obtained from vaginal swabs. As seen in Fig. 1, we had an average of 17,427 sample counts per sample (range: 10435 – 30821) with no significant differences in sample counts between sampling methods (Figure 1C). The ten most abundant bacterial genera in the lower FGT were Porphyromonas, Fusobacterium, Acholeplasma, Peptoniphilus, Catonella, Prevotella, Campylobacter, Mobiluncus, Clostridium, and Dialister (Figure 1D). Principal Coordinate analysis (PCoA) using weighted UniFrac distance metric showed that the relative abundance of bacterial taxa was comparable across sampling methods within animals (Figure 1E). Compositional differences in bacterial taxa in the younger female relative to the older females raise the possibility of age-dependent changes in microbial diversity, which should be investigated in larger studies.
DISCUSSION
Here we report that CVL is as effective as swabs in providing a comprehensive overview of the vaginal microbiome. These findings are consistent with a recent study in humans showing that sampling by flocked swabs, plastic spatula, and cervical brush yielded comparable vaginal microbiota composition(8).
Unlike humans, however, the rhesus microbiome is highly polymicrobial with an average representation of twelve bacterial genera in each animal. The observation that both swabs and CVL samples capture this polymicrobial bacterial composition has important practical implications for NHP studies. The data indicate that swabs can be prioritized for humoral assays and that CVL sampling provides an analogous profile of microbial composition. It is noteworthy that cellular CVL fraction was also suitable for determining the microbiology of the FGT despite being highly enriched for host DNA. This information supports the adoption of FGT sampling via CVL in HIV vaccine/prevention studies.
ACKNOWLEDGEMENTS
The authors acknowledge the Primate Services Unit staff at the CNPRC including Miles Christensen and Wilhelm Von Morgenland for their contributions that have resulted in this report. This study was supported by the California National Primate Research Center base grant P51 OD011107, and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under award number K01OD023034 and R03 AI13879 (to SSI) and the National Institute of Diabetes And Digestive And Kidney Diseases of the NIH under award number K01DK110264 (to RR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, Dong M, Chen Y, Ismail N, Ndung’u T, Ghebremichael MS, Wesemann DR, Mitchell C, Dong KL, Huttenhower C, Walker BD, Virgin HW, Kwon DS. Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women. Immunity. 2017;46(1):29–37. 10.1016/j.immuni.2016.12.013 PubMed PMID: ; PMCID: PMC5270628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev 2013;37(5):762–92. doi: 10.1111/1574-6976.12029 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 3.Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, Padavattan N, Ismail N, Moodley A, Sabatini ME, Ghebremichael MS, Nusbaum C, Huttenhower C, Virgin HW, Ndung’u T, Dong KL, Walker BD, Fichorova RN, Kwon DS. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42(5):965–76. 10.1016/j.immuni.2015.04.019 PubMed PMID: ; PMCID: PMC4461369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spear G, Rothaeulser K, Fritts L, Gillevet PM, Miller CJ. In captive rhesus macaques, cervicovaginal inflammation is common but not associated with the stable polymicrobial microbiome. PLoS One. 2012;7(12):e52992. doi: 10.1371/journal.pone.0052992 PubMed PMID: ; PMCID: PMC3532444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spear GT, Gilbert D, Sikaroodi M, Doyle L, Green L, Gillevet PM, Landay AL, Veazey RS. Identification of rhesus macaque genital microbiota by 16S pyrosequencing shows similarities to human bacterial vaginosis: implications for use as an animal model for HIV vaginal infection. AIDS Res Hum Retroviruses. 2010;26(2):193–200. doi: 10.1089/aid.2009.0166 PubMed PMID: ; PMCID: PMC2835387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer SS, Sabula MJ, Mehta CC, Haddad LB, Brown NL, Amara RR, Ofotokun I, Sheth AN. Characteristics of HIV target CD4 T cells collected using different sampling methods from the genital tract of HIV seronegative women. PLoS One. 2017;12(6):e0178193. doi: 10.1371/journal.pone.0178193 PubMed PMID: ; PMCID: PMC5453484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. 10.1128/AEM.71.12.8228-8235.2005 PubMed PMID: ; PMCID: PMC1317376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virtanen S, Kalliala I, Nieminen P, Salonen A. Comparative analysis of vaginal microbiota sampling using 16S rRNA gene analysis. PLoS One. 2017;12(7):e0181477 10.1371/journal.pone.0181477 PubMed PMID: ; PMCID: PMC5517051. [DOI] [PMC free article] [PubMed] [Google Scholar]


