Abstract
Damage to DNA via dissociative electron attachment has been well-studied in both the gas and condensed phases; however, understanding this process in bulk solution at a fundamental level is still a challenge. Here, we use a picosecond pulse of a high energy electron beam to generate electrons in liquid diethylene glycol and observe the electron attachment dynamics to ribothymidine at different stages of electron relaxation. Our transient spectroscopic results reveal that the quasi-free electron with energy near the conduction band effectively attaches to ribothymidine leading to a new absorbing species that is characterized in the UV-visible region. This species exhibits a nearly concentration-independent decay with a time constant of ~350 ps. From time-resolved studies under different conditions, combined with data analysis and theoretical calculations, we assign this intermediate to an excited anion radical that undergoes N1-C1′ glycosidic bond dissociation rather than relaxation to its ground state.
Radiation-induced low-energy electrons in solution are implicated in DNA damage, but their relaxation dynamics are not well understood. Here the authors observe how quasi-free electrons dissociate glycosidic bonds via an excited nucleoside anion radical, whereas solvated electrons reside on the nucleoside as a relatively stable anion radical.
Introduction
Radiation-induced cellular DNA damage stems not only from the impact (i.e. direct effect) of primary high-energy photons and charged particles, but also from secondary species (excited molecules, free radicals, and free electrons) that are produced via radiolysis of cell components along the radiation tracks1,2. Secondary electrons are ubiquitous in an irradiated medium with an estimated quantity of ~4 × 104 electrons per 1 MeV energy deposited3. They cause cascades of additional ionizations and excitations through inelastic scattering with molecules. As a result, low-energy electrons (LEEs) are generated with an excess kinetic energy of 0–20 eV4.
DNA strand breaks, especially double strand breaks (DSBs), are the most important DNA damage that has been shown to lead to cell death and neoplastic transformation1,5. It is known that fully solvated electrons (esol-) are ineffective at triggering DNA bond cleavage because they generally reside on biomolecules as stable anions6. For this reason, the conventional notion of electron-induced damage to the genome is mainly due to those electrons with sufficient energy to ionize or excite DNA, thereby leading to the formation of electron-loss radicals (holes) and excited states that cause subsequent molecular fragmentation7. In 2000 and onwards, the experimental observations from Sanche and coworkers showed that LEEs were able to cause single strand breaks (SSBs), as well as DSBs via dissociative electron attachment (DEA)8,9. This observation motivated a great number of mechanistic studies on the interaction of LEEs with DNA and its components in both the gas and condensed phases10–15. The low-energy resonance features in the yield of DSBs, SSBs, and anions produced by the impact of LEEs on model pyrimidine bases suggested that the initial step involves electron capture into the unoccupied molecular orbitals that are above the lowest unoccupied molecular orbitals (LUMOs) of the parent nucleobase, creating excited transient negative ions (TNIs*). Once the TNIs* are formed, they are shown to decay very rapidly leading either to a SSB via phosphate-sugar C–O σ bond cleavage12,13 or result in unaltered base release via N1–C1′ glycosidic bond breakage14,15. Studies of DEA using various DNA models (monomers, oligomers with defined sequences, plasmid DNA) were often carried out under vacuum conditions; these experiments were limited to gas phase and condensed phase or micro-hydrated molecular targets10–15.
In a polar medium (e.g. water), as shown in Fig. 1, LEEs successively lose energy to become quasi-free electrons (eqf-) and they can undergo multistep solvation prior to their complete localization as esol-2,16,17. The transition from eqf- to esol- is accompanied by the appearance of a strong optical absorption as the electron acquires a stable quantum state of binding energy, which was evidenced by time-resolved techniques, typically using a short pulse of high-energy electrons or a laser beam16–18. From the viewpoint of the action of LEEs, it is appropriate to suggest that a thorough understanding of the role played by short-lived non-equilibrated electrons would lead to a clearer picture of the basic mechanisms underlying the biological consequences of radiation. Therefore, a detailed knowledge of electron attachment to DNA/RNA in solution leading to the formation of the TNI* and the subsequent pathways of reactions that the TNI* undergoes, are of fundamental importance. However, these studies, even at a monomeric DNA-subunit (e.g., nucleosides, nucleotides) level, have been lacking. This may be due to challenges encountered in femtosecond laser spectroscopic investigations on the formation of TNI* and its reaction channels19. In contrast, the accelerator technique delivers a high-energy electron pulse to the solvent, and hence generates LEEs in accord with those in radiation biology and allows us to investigate the chemistry induced by radiation-produced electrons in liquids19.
Fig. 1.
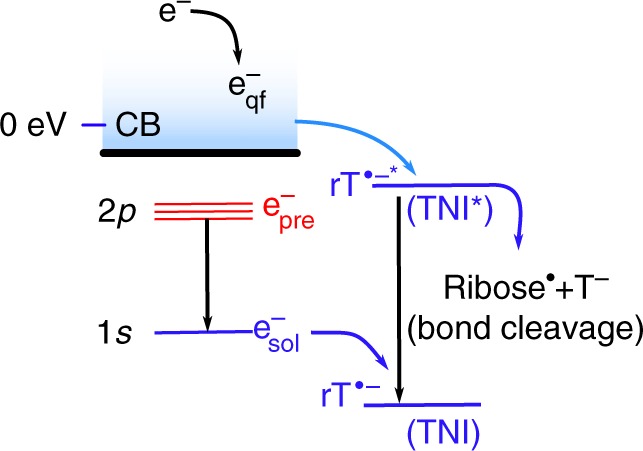
A schematic diagram of energy level showing different states of electrons during trapping and relaxation. These processes take place in a polar medium following ionizing radiation in the presence of ribothymidine (rT). An excess electron in the conduction band (CB), representing a quasi-free electron (eqf-), eventually becomes trapped (esol-) in the solvent cage. The excited state of esol- is considered as a "presolvated" electron, epre-. Electrons captured by solute molecules produce transient negative ions (TNI or rT•‒). The TNI in its excited state (TNI*) can either liberate the excess energy to the solvent (relaxation) or undergo bond breaking (dissociation)
Unfortunately, under ambient conditions, as the solvation of LEEs in water is fast (≤1 ps), and the limitation of current high-energy electron pulse (7 ps) radiolysis system19 prevents investigation of the complete time resolution of the epre- solvation versus its attachment processes. Relaxation of the electron (i.e. from eqf- to epre- and ultimately to esol-, Fig. 1) can be viewed as a multistep transition from the delocalized conduction band with p-like excited states to s-like ground states2,16,20; however, experimental characterization of the specific state of electron that is required for the DEA processes still remains elusive. In addition, formation of the excited states of DNA anion radicals via electron attachment has been suggested but has never been observed1,19.
It is known that the time of solvation of electrons in alcohols is of the order of several ps18,20. Here, we observe that electron solvation events in diethylene glycol (DEG) are relatively slow and occur on the order of tens of picoseconds. As the value of the dielectric constant (εr = 31.69) of DEG is much closer to that of a biological cell21, it allows us to investigate the reactions of electrons that are more relevant to biological system. Also, DNA can retain its native double-stranded structure and biological activity in glycol solutions22. We solely find that ribothymidine (rT), a DNA/RNA model, can be sufficiently dissolved in DEG (up to 0.5 M) to scavenge electrons on a short time scale. We conduct picosecond pulse radiolysis studies of rT solutions in DEG at different concentrations (0–0.5 M) under ambient conditions, that allow us to observe real-time dynamics of the electron attachment to DNA-components at certain stages of electron relaxation. Our pulse radiolysis technique23 is based on a picosecond laser-triggered electron accelerator coupled with transient UV–Vis/IR absorption spectroscopy (Supplementary Figure 1). The key transient species (epre-, esol-, and TNI* of rT•‒*) can be directly observed by the probe light covering a broad wavelength range from 370 to 1100 nm. In addition, theoretical calculations using density functional theory (DFT) show that rT•‒* undergoes N1–C1′ glycosidic bond dissociation rather than relaxation to its ground state.
Results
Electron solvation in liquid DEG
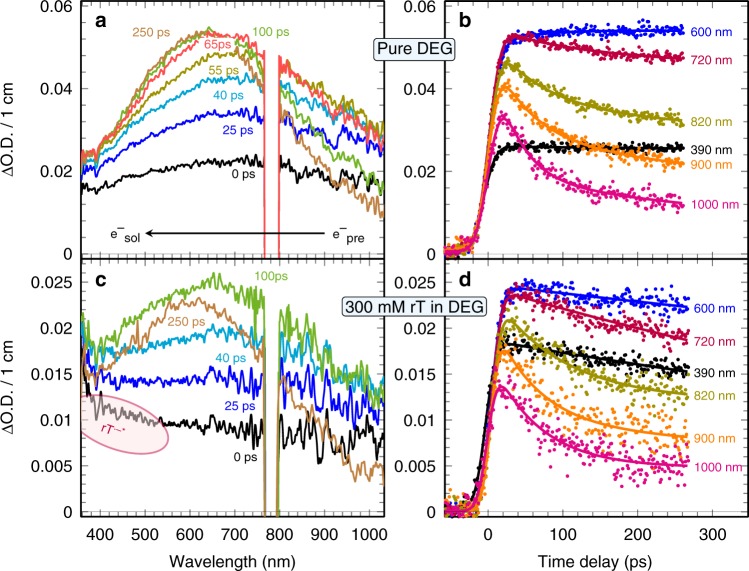
The transient absorption spectra of neat DEG is shown in Fig. 2a. A significant fraction of electrons formed in DEG immediately after the electron pulse have undergone relaxation. A transient feature absorbing above 900 nm rapidly diminishes, and a broad intense signal with a maximum at around 750 nm undergoes a continuous blue shift of ca.120 nm, accompanied with a growing amplitude of about hundreds of picoseconds (e.g. 250 ps, Fig. 2a). At the end of the spectral evolution (Fig. 2a), a broad and featureless absorption band builds up with a peak at 630 nm. Based on previous studies in various alcohols24, this band is assigned to the spectrum of esol-. Quenching of the signals by adding acetone (an electron scavenger) confirms this assignment and nanosecond pulse radiolysis measurements indicated that the lifetime of esol- in neat DEG is around 5 μs. Alcohol radicals are formed by ionization, but they absorb only in the UV region below 300 nm. As a result, the time-dependent spectral changes in Fig. 2a clearly show that the electron solvation essentially consists of at least two distinct states, one absorbing in the near-infrared, and one in the visible range.
Fig. 2.
Transient absorption profiles of pure diethylene glycol and ribothymidine solutions under ambient conditions. Spectra (a, c) and kinetics (b, d) for neat DEG and 0.3 M ribothymidine (rT) solutions, respectively. The region at 780 nm is filtered out. The duration of the electron pulse is ~7 ps. The dose per electron pulse is 25.3 Gy
Figure 2b presents kinetic traces of the transient electrons in DEG at various wavelengths (390–1000 nm) with a non-monotonic kinetics and with no isosbestic point. Figure 2b clearly shows that the signals at 390 and 600 nm rise fast, then remain almost constant for the next hundreds of ps, and the one at 900–1000 nm follows a continuous decrease. The overall behavior of transient spectra and kinetics show a stepwise transition between the two above-mentioned states (τ1) along with simultaneous blue shift of visible state (τ2). To gain further insights into the dynamics, the best multi-exponential global fits to transients at a variety of wavelengths find two components, a fast component with a time constant of τ1 = (45 ± 15) ps and a slow component with τ2 = (80 ± 30) ps. Our results are in agreement with the previously reported spectral behaviors of electrons in liquid water25,26, methanol27,28, ethylene glycol29, and polyols30 observed by femtosecond laser spectroscopy.
In alcohols, the formation time of esol- was reported to be much slower than in water and it is determined by many factors, such as the size of alcohol, the number of the hydroxyl group and dielectric relaxation time for molecular rotation. Our model that uses multi-exponential global fits to account for transients involved in electron solvation processes, agrees well with the hybrid model proposed by Pépin et al.25,27 and Barbara et al.26,28. According to this model and theoretical simulations31–33, the faster relaxation component, τ1, can be assigned to the transition of the weakly bound p-like states, and the slower continuously shifting component, τ2, is attributed to p → s radiationless decay or cooling of a vibrationally excited s-state. Recent liquid-jet photoelectron spectroscopy results supported the latter view34,35. For instance, angular-resolved photoemission measurements showed the photoelectrons generated from the state of the slow component is associated with an isotropic character, revealing the existence of the “hot” ground state35. Therefore, our results on the precursors of esol- and their equilibration process in DEG represent a starting point for further investigation of the electron transfer process to DNA constituents.
Electron attachment to rT leading to TNI* formation
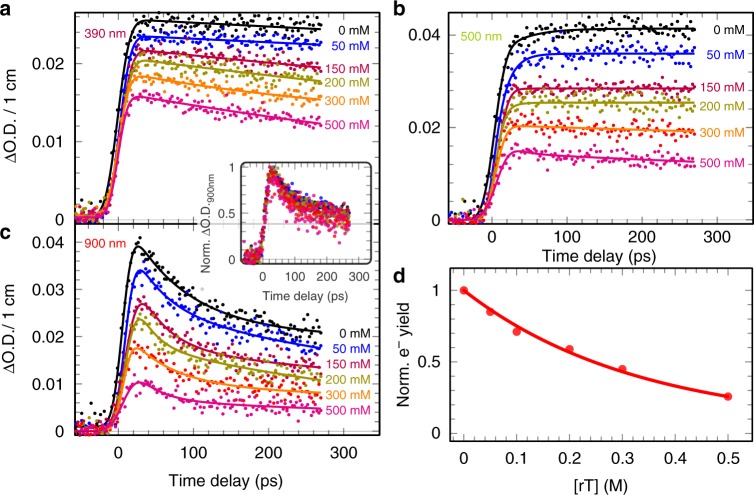
To investigate the electron attachment to rT and the consequent formation of rT•‒*, picosecond pulse radiolysis studies were performed in rT solutions (50–500 mM) in DEG. In these solutions, direct ionization or excitation of solute itself is not significant. As an example, the transient spectra and the kinetics at different wavelengths are reported for the concentration of 300 mM in Fig. 2c, d, and the effect of the rT concentration on the kinetics are shown in Fig. 3. Rapid electron capture by rT generates a new transient signal that is immediately and clearly visualized in the UV–visible regions shown in Fig. 2c. In contrast to the slight increase in neat DEG, the transient kinetics in the UV–visible region (370–600 nm) shown in Fig. 2d and Fig. 3a, b of rT solutions display an obvious decay. From Fig. 3c, d, we observe a substantial decrease of the initial near-infrared absorbance that correlates exponentially with increasing rT concentrations. These results clearly show that rT has scavenged a significant fraction of the electrons prior to their being trapped in DEG. For instance, about 12% and as high as 75% of electrons are captured by rT molecules at the concentration of 0.05 and 0.5 M, respectively. Analyses of normalized kinetics at the infrared region from 720 to 1000 nm, at which the absorptions are only associated with epre-, are found to be nearly identical (Fig. 3d, inset) with those observed in neat DEG (Fig. 2b). Also, the global fitting for the transient in each rT solution at the higher wavelength (>720 nm) shows that the characteristic times of two components (τ1 and τ2) remain almost unchanged. Therefore, these results establish that even such a significant extent of electron scavenging takes place before electron localization in pre-existing traps, the presence of rT does not affect significantly the subsequent electron solvation process in DEG (Fig. 3); also, rT does not react with epre- on the time scale of hundreds of ps. Based upon our assignments of the states of epre- and esol-, we conclude that the electrons captured by rT within the pulse duration (≤7 ps) are most likely attributed to eqf- at or above the conduction band. Because energy levels of trapped electrons in a solvent strongly correlate with time of electron solvation36–38, it is emphasized that a small but non-negligible energy barrier prevents the ultrafast attachment by electrons lying at lower energy trapping sites, such as p-like states (component τ1), vibrationally excited s-like state (component τ1), and ground s-like state (esol¯). Indeed, the complete relaxation of electrons starts from delocalization of excess electrons possessing a high mobility (>1 cm2 V−1 s−1)20. In water and alcohols, the vertical binding energy (VDE) of a conduction band electron is much lower than that of its weakly bound states and ground states37–40. Moreover, first-principle molecular dynamics simulations predicted that excess electrons attach rapidly (<15 fs) to solvated DNA nucleotides41, which supports our experimental results. Therefore, based on these studies and on our results (Figs. 2 and 3), the higher chemical reactivity of eqf- relative to that of trapped electrons is justified. In addition, our findings establish the quantum states of electrons that are involved in formation of TNI* via ultrafast electron attachment to DNA-components in solution.
Fig. 3.
Transient absorption kinetics at various ribothymidine concentrations. Kinetics at wavelengths: a 390 nm, b 500 nm, and c 900 nm. The inset shows the normalized kinetics at 900 nm at different concentration of ribothymidine (rT). Electron solvation is shown to be complete within about 300 ps and the geminate recombination between the electron and hole can be neglected at this timescale. d Relative electron yield in the presence of rT
As described above, the presence of rT has only changed the initial absorbance of epre- and the observed IR spectra of epre- are identical with that in DEG. Consequently, the transient profiles of rT intermediates were obtained as shown in Fig. 4, via a combination of a multivariate curve resolution alternating least squares (MCR-ALS) analysis of a data matrix (Supplementary Figure 2) previously described42 and via simply subtracting the epre- absorption in neat DEG (Supplementary Figure 3). In studied wavelength range, the spectra of this species are characterized as a mono-peak absorption band extending to the UV (Fig. 4a) and it does not evolve with the delay time (Supplementary Figure 3). Figure 4a inset also showed that this species undergoes a decay that is nearly independent of the concentration. The lifetime of this species is estimated to be ~350 ps based on the lineally fitting of the kinetic in logarithm, as well as the half-life of TNI* shown in Fig. 4b, which agrees with previously measured rates of charge-induced dissociation43.
Fig. 4.
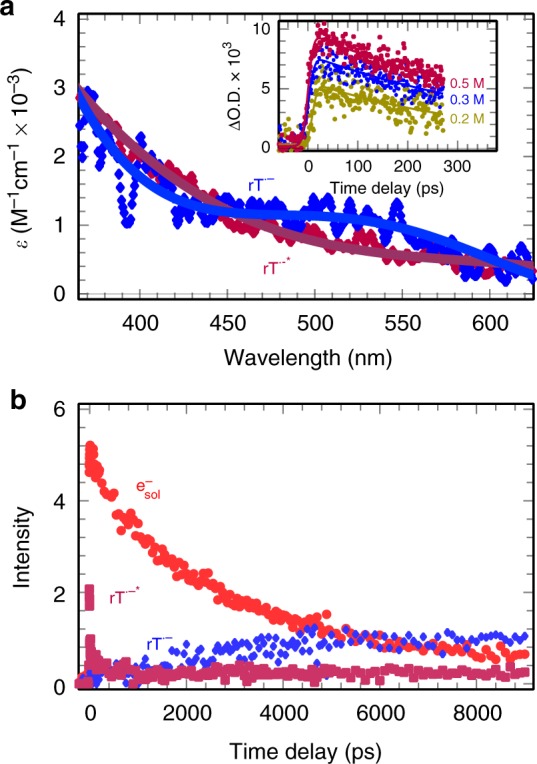
Deconvolution of absorbing species in a data matrix. a Spectra of excited and ground states of ribothymidine anion radicals, rT•‒* and rT•‒, respectively. The inset shows the kinetics of rT•‒* at different concentrations. b Deconvolution of transient data using a multivariate curve resolution alternating least squares (MCR-ALS) method representing the kinetics of rT•‒*, rT•‒, and the solvated electron (esol¯)
In the liquid phase, electron attachment to rT results in the formation of excited rT•‒* (i.e., TNI*) or ground state, rT•‒, i.e., TNI. rT•‒* will subsequently, either dissociate to a neutral radical (R•) and an anion when the energy is accessible or relax to a stable anion radical by liberating energy to the solvent. This latter species, rT•‒, is essentially, identical in nature with that from the reaction of the fully esol¯ (see Fig. 1). It is less likely that the transient kinetics correspond to the decay of fragment radicals R•. This is because in DEG (viscosity = 35.7 cP; 25 °C), R• should react with other radicals through nearly diffusion-limited reactions on timescale of tens of nanoseconds. In addition, the decay rate should be affected by the concentration of R•. More importantly, the initial absorbance of the new species at various concentrations is linearly correlated with the number of electrons captured (Supplementary Figure 4). After ruling out this possibility, the key question now is whether it corresponds to the excited anion radicals or fully solvated anion radicals or both. To answer this question, the MCR-ALS analysis of a data matrix showed several important features of species involved as displayed in Fig. 4: (i) only two absorbing species (e‒ and rT•‒*) exist at the early time. rT•‒* is formed within the pulse and not after the electron pulse (ii). rT•‒* decays in a few 100 ps and the decay of the electron during this time is almost negligible (Fig. 4b). (iii) The decay of the rT•‒* does not form the rT•‒ because the formation of the latter only correlates with the decay of esol- at longer time (Fig. 4b). Besides, to compare the spectral difference (the band shape, lifetime, and extinction coefficient) between rT•‒* and rT•‒, we performed additional measurements of DEG solution at low concentrations (1–10 mM), in which eqf¯ and epre- are not scavenged by rT at long range and all of them are converted into rT•‒ (Fig. 1). The resulting anion radicals rT•‒ at microsecond timescale show a distinct transient spectrum (blue curve in Fig. 4a) and remain stable over a long period of time (lifetime >10 μs, see Supplementary Figure 5). These results conclude that the transient signals are attributed to the excited anion radical rT•‒* formed from eqf¯ attachment to rT in liquid DEG.
In contrast to the results obtained in DEG solutions, studies using aqueous rT did not provide any evidence of the bond breaking. All TNIs* relaxed to rT•‒ (Fig. 1), which is in the agreement with our previous work19. For a given rT concentration, e.g., 0.5 M, the scavenging time of epre- is about 400 fs both in water and in DEG as the rate constant of epre- scavenging by rT is found to be similar, ~5 × 1012 M−1 s−1. However, as the solvation dynamics in water is much faster, the time for excess electrons reaching into the pre-existing traps of liquid water is on the order of tens of fs and the lifetime of p-like states of electron was around 300 fs. Thus, rT in liquid water cannot react with the higher electronic states of electrons, such as p-like states or conduction states, which, in turn, do not offer enough energy for the bond rupture of TNIs*. Thus, our results clearly point out the energy states of a single electron and pre-existing traps in the solvent medium are the decisive factor for the occurrence of TNIs* fragmentation.
LEEs-induced decomposition of thymidine was first studied by analysis of irradiated products at the molecular level in the condensed phase14. These experiments showed the effective excision of the thymine base from low-energy electron-irradiated DNA oligomers of defined sequences through N1–C1′ glycosidic bond cleavage. In the mechanistic study, Ptasinska et al.15 suggested that the resonant localization of an electron with an energy as low as 1.2 eV on the sugar moiety in the gas-phase can cause bond cleavage. Kočišek and co-workers44 recently provided better insight into the role of water played in DEA using a “bottom-up” approach in a molecular beam, by progressively micro-hydrating the target molecule. This study highlights the suppression of dissociation of N1–C1′ glycosidic bond by increasing the hydration degree of thymine and uracil. Thus, the decay of rT•‒* observed in our time-resolved experiments that we have assigned to a dissociative process could be associated with the dynamics of glycosidic bond cleavage14.
Modeling of excited TNI surfaces leading to N1–C1′ bond breakage
It is well-known that LEEs on interacting with a molecule create TNI resonances which are equivalent to vertical excited states of the electron adduct of the parent molecule45–49. Based on this understanding, calculations of the transition energies to vertical excited states of a TNI can predict the specific resonance energies available for direct capture of LEEs. In this work, the transition energies of TNI of rT in DEG (εr = 31.69) are calculated using the time-dependent DFT (TD-DFT) implemented in Gaussian 1650. The complete methodology is abbreviated as TD-ωB97XD-PCM/6-31G**. Use of a compact basis set (6–31G**) in these calculations avoids mixing of resonances with the continuum46–48,51.
From the nature of the TNI potential energy surface (PES) (2πTNI, Fig. 5), we see that as the N1–C1’ bond elongates the energy of ground state of the TNI (2πTNI) surface increases until it crosses the dissociative 2σ* surface with a barrier of 1.6 eV. The energy of the first excited state 2π → 2πσ* (2πσ* surface) also increases as N1–C1′ bond elongates and at 1.8 Å it crosses the dissociative 2σ* surface having a barrier of ca. 1 eV. We also optimized the 2πσ* excited state (The 2πσ*OPT was not fully optimized because during optimization this excited state switches to another excited state after few cycles of the optimization steps. Thus, we use the energy of 2πσ*OPT state just before the switch to the other excited state.) designated as 2πσ*OPT in Fig. 5. As expected, the adiabatic state, 2πσ*OPT, lies lower than the vertical 2πσ* surface and after surpassing a small barrier of ca. 0.6 eV, the N1–C1′ bond dissociates. The light blue solid line in Fig. 5, denotes a proposed barrier-free dissociation path which occurs on capture of a quasi-free electron into the vertical πσ-MO of rT (2πσ*) which upon extension of the N1–C1′ bond, relaxes from the vertical to the adiabatic surface.
Fig. 5.
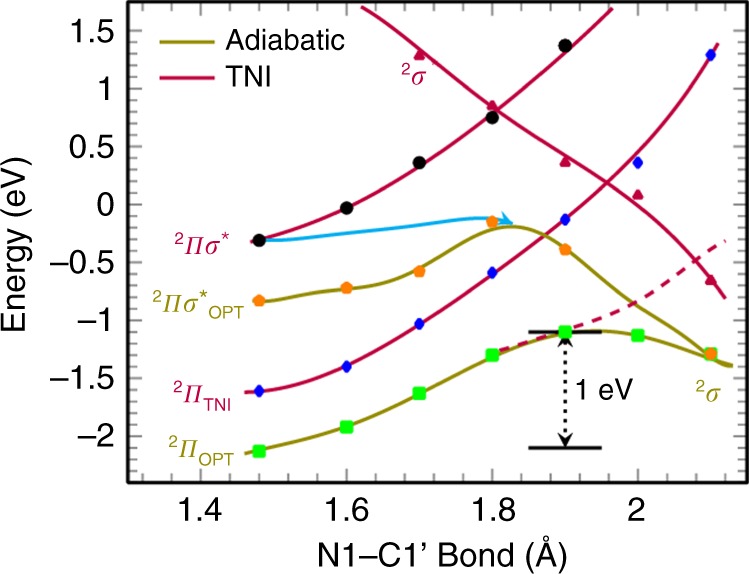
Potential energy surface profiles for N1–C1′ bond cleavage. Shown for the ground and vertical excited states of rT anions (TNI*). The energies were scaled by adding −1.6 eV to the actual calculated values (Supplementary Figure 8) to match with the theoretical adiabatic electron affinity (2.1 eV) of rT. The light blue solid line is the proposed path leading to barrierless fast N1–C1′ bond dissociation
The ground state adiabatic PES of rT anion radical (2πOPT, Fig. 5) shows that the N1–C1′ bond dissociation involves a substantial barrier ca. 1 eV as reported earlier45. From the overall nature of the PESs, we inferred that the energy of the lowest excited state of the TNI is located at −0.3 eV (2πσ*) and provides the most likely path for immediate dissociation of rT via electron attachment. To mimic the experiment, we scaled the energy in Fig. 5 by adding −1.6 eV to the calculated values for matching the adiabatic electron affinity (AEA) (2.1 eV) of rT that were calculated using the ωB97XD-PCM/6-31++G** method with actual energy values presented in the supporting information (Supplementary Figures 6–9). From Fig. 5, it is evident that the vertical 2πσ* excited state of TNI has energy of −0.3 eV which lies within the conduction band energy range. An overview of the energies of the electron and TNI of rT in DEG is shown in Fig. 1 and Supplementary Figure 9. Thus, electrons generated in the conduction band should be efficiently captured into the excited state πσ*-MO of the rT TNI and proceed via gradual relaxation of the structure on bond elongation to a barrier-free N1–C1′ glycosidic bond cleavage leading to thymine release.
Discussion
Our spectroscopic observations in DEG establish that the quasi-free electrons form two localized electron-solvent configuration states (in the infrared region and in the visible region within the timescale of the electron pulse (<7 ps)); the former is characterized as a p-like state and the latter is assigned to a vibrationally hot ground state which gradually relaxes to form a solvated electron esol-. In the presence of rT, our results show that dissociative electron transfer occurs only by the quasi-free electrons at or above the conduction band rather than via epre- and esol-. The resulting excited rT anion radical rT•‒* observed on a picosecond timescale has been fully characterized, and it exhibits a spectrum that is different from the spectrum of the stable anion radical rT•‒ which is observed on the microsecond timescale in dilute DEG solutions at room temperature. The combination of time-resolved results and DFT calculations establishes that the observed transients rT•‒* can be attributed to the excited state πσ*-MO of the TNI of rT, and its dissociation proceeds via gradual relaxation of the structure on bond elongation through a barrier-free N1–C1′ glycosidic bond cleavage. Our results further imply the generation of biomolecular damage does not necessarily require electrons carrying kinetic energy. In cellular systems the water molecules have inherently long relaxation times. Conduction band electrons (eqf-) formed in cells should have longer lifetimes than those found in water or in DEG; as a result, these longer-lived eqf- would contribute to biomolecular damage. Thus, the insights gained in our present study could pave the way to directly investigate the long-standing mystery of electron-driven reactions in radiation chemistry and biology.
Methods
Pulse radiolysis experiment
The chemical compounds (rT and DEG; purity, >99%) were purchased from Sigma-Aldrich and used without further purification. The 7 ps pulse radiolysis coupled with transient absorption measurements were performed at the electron facility LINAC (Tokyo University) coupled to a transient absorption broadband probe spectroscopy (380–1050 nm) instrument. Additional time-resolved radiolysis results were also carried out at ELYSE facility (Paris-Saclay University) for comparison. The experimental data matrices were analyzed by a MCR-ALS approach as previously described52. Details about experimental apparatus, methodologies, and data analysis are provided in Supplementary Notes 1 and 2.
TD-DFT calculations
In this study, the ωB97XD density functional and 6–31G** basis set were used and the effect of bulk solvent with dielectric constant of DEG (ε = 31.69) was incorporated via the use of the integral equation formalism polarized continuum model (IEF-PCM). Details about the TD-DFT calculations are provided in Supplementary Note 3.
Supplementary Information
Acknowledgements
This work was partially supported by Grant-in-Aid for Scientific Research Nos. 17F17085, 15K21721, 15H04243, and 18H03918) from Japan Society for the Promotion of Science (JSPS). The authors thank Dr. Mitsumasa Taguchi (Takasaki Advanced Radiation Research Institute, National Institute for Quantum and Radiological Science and Technology, QST) for γ-ray irradiation as well as the Shared-Use Program of QST. J.M. thanks the Japan Society for the Promotion of Science for a Fellowship. A.K., M.D.S., and A.A. thank the National Cancer Institute of the National Institutes of Health (Grant R01CA045424) for support. A.A. is also grateful to Research Excellence Fund (REF) from Center for Biomedical Research (CBR) at Oakland University for support. It is our pleasure to thank Prof. M. Uesaka and Mr. T. Ueda (The University of Tokyo) for providing support with the accelerator experiment. We thank Dr. Tamura (Kyoto University) for providing support with the HPLC measurements. The authors are grateful to Prof. David Becker (Chemistry Department, Oakland University) for copy-editing the revised manuscript.
Author contributions
This work resulted by the collaboration among J.M., M.M., S.S., and A.A.; J.M., M.M., and S.S. designed the experimental part of the project; J.M., Y.M., and S.Y. performed key experiments; T.S., S.A.D., S.S., and M.M. performed additional necessary experiments; A.K. and M.D.S. performed TD-DFT calculations and analyzed data; J.M., S.A.D., and M.M. analyzed the experimental data; A.A., J.M., S.S., A.K., M.D.S., and M.M. wrote the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contributions to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jun Ma, Email: ma.jun.26m@st.kyoto-u.ac.jp.
Shu Seki, Email: seki@moleng.kyoto-u.ac.jp.
Mehran Mostafavi, Email: mehran.mostafavi@u-psud.fr.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-08005-z.
References
- 1.von Sonntag, C. Free-Radical-Induced DNA Damage and Its Repair: A Chemical Perspective (Springer, Berlin, Heidelberg, 2006).
- 2.Alizadeh E, Sanche L. Precursors of solvated electrons in radiobiological physics and chemistry. Chem. Rev. 2012;112:5578–5602. doi: 10.1021/cr300063r. [DOI] [PubMed] [Google Scholar]
- 3.Cobut V, et al. Monte Carlo simulation of fast electron and proton tracks in liquid water. I. Physical and physicochemical aspects. Radiat. Phys. Chem. 1998;51:229–243. doi: 10.1016/S0969-806X(97)00096-0. [DOI] [Google Scholar]
- 4.Pimblott SM, LaVerne JA. Production of low-energy electrons by ionizing radiation. Radiat. Phys. Chem. 2007;76:1244–1247. doi: 10.1016/j.radphyschem.2007.02.012. [DOI] [Google Scholar]
- 5.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 6.Nabben FJ, Karman JP, Loman H. Inactivation of biologically active DNA by hydrated electrons. Int. J. Radiat. Biol. 1982;42:23–30. doi: 10.1080/09553008214550881. [DOI] [PubMed] [Google Scholar]
- 7.Becker, D., Adhikary, A. & Sevilla, M. D. in Mechanisms of Radiation-induced DNA Damage: Direct Effects. Recent Trends in Radiation Chemistry (eds. Wishart, J. F. & Rao, B. S. M.) 21–58 (World Scientific Publishing Company, 2010).
- 8.Boudaiffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science. 2000;287:1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 9.Martin F, et al. DNA strand breaks induced by 0–4 eV electrons: the role of shape resonances. Phys. Rev. Lett. 2004;93:068101. doi: 10.1103/PhysRevLett.93.068101. [DOI] [PubMed] [Google Scholar]
- 10.Ptasinska S, Denifl S, Scheier P, Illenberger E, Märk TD. Bond- and site-selective loss of H atoms from nucleobases by very-low-energy electrons (<3 eV) Angew. Chem. Int. Ed. Engl. 2005;44:6941–6943. doi: 10.1002/anie.200502040. [DOI] [PubMed] [Google Scholar]
- 11.Yandell MA, King SB, Neumark DM. Time-resolved radiation chemistry: photoelectron imaging of transient negative ions of nucleobases. J. Am. Chem. Soc. 2013;135:2128–2131. doi: 10.1021/ja312414y. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Leszczynski J, Schaefer HF., III. Interactions of electrons with bare and hydrated biomolecules: From nucleic acid bases to DNA segments. Chem. Rev. 2012;112:5603–5640. doi: 10.1021/cr3000219. [DOI] [PubMed] [Google Scholar]
- 13.Kopyra J. Low energy electron attachment to the nucleotide deoxycytidine monophosphate: direct evidence for the molecular mechanisms of electron-induced DNA strand breaks. Phys. Chem. Chem. Phys. 2012;14:8287–8289. doi: 10.1039/c2cp40847c. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Y, Cloutier P, Hunting DJ, Wagner JR, Sanche L. Glycosidic bond cleavage of thymidine by low-energy electrons. J. Am. Chem. Soc. 2004;126:1002–1003. doi: 10.1021/ja0388562. [DOI] [PubMed] [Google Scholar]
- 15.Ptasińska S, et al. Decomposition of thymidine by low-energy electrons: Implications for the molecular mechanisms of single-strand breaks in DNA. Angew. Chem. Int. Ed. 2006;45:1893–1896. doi: 10.1002/anie.200503930. [DOI] [PubMed] [Google Scholar]
- 16.Migus A, Gauduel Y, Martin JL, Antonetti A. Excess electrons in liquid water: first evidence of a prehydrated state with femtosecond lifetime. Phys. Rev. Lett. 1987;58:1559–1562. doi: 10.1103/PhysRevLett.58.1559. [DOI] [PubMed] [Google Scholar]
- 17.Shkrob, I. A. In The Structure and Dynamics of Solvated Electrons.Recent Trends in Radiation Chemistry (eds. Wishart, J. F. & Rao, B. S. M.) 21–58 (World Scientific Publishing Company, 2010).
- 18.Chase WJ, Hunt JW. Solvation time of the electron in polar liquids. Water Alcohols J. Phys. Chem. 1975;79:2835–2845. [Google Scholar]
- 19.Ma J, et al. Reactivity of prehydrated electrons toward nucleobases and nucleotides in aqueous solution. Sci. Adv. 2017;3:1–7. doi: 10.1126/sciadv.1701669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenney - Wallace GA, Jonah CD. Picosecond spectroscopy and solvation clusters. The dynamics of localizing electrons in polar fluids. J. Phys. Chem. 1982;86:2572–2586. doi: 10.1021/j100211a007. [DOI] [Google Scholar]
- 21.Goncharenko AV, Chang YC. Effective dielectric properties of biological cells: generalization of the spectral density function approach. J. Phys. Chem. B. 2009;113:9924–9931. doi: 10.1021/jp900703a. [DOI] [PubMed] [Google Scholar]
- 22.Eliasson R, Hammarsten E, Lindahl T, Björk I, Laurent TC. The stability of deoxyribonucleic acid in glycol solution. Biochim. Biophys. Acta. 1963;68:234–239. doi: 10.1016/0926-6550(63)90434-1. [DOI] [Google Scholar]
- 23.Muroya Y, Lin MZ, Iijima H, Ueda T, Katsumura Y. Current status of the ultra-fast pulse radiolysis system at NERL, the University of Tokyo. Res. Chem. Intermed. 2005;31:261–272. doi: 10.1163/1568567053146850. [DOI] [Google Scholar]
- 24.Jou FJ, Freeman GR. Band resolution of optical spectra of solvated electrons in water, alcohols, and tetrahydrofuran. Can. J. Chem. 1979;57:591–597. doi: 10.1139/v79-095. [DOI] [Google Scholar]
- 25.Pépin C, Goulet T, Houde D, Jay-Gerin JP. Observation of a continuous spectral shift in the solvation kinetics of electrons in neat liquid deuterated water. J. Phys. Chem. A. 1997;101:4351–4360. doi: 10.1021/jp970354l. [DOI] [Google Scholar]
- 26.Yokoyama K, Silva C, Son DH, Walhout PK, Barbara PF. Detailed Investigation of the femtosecond pump-probe dpectroscopy of the hydrated electron. J. Phys. Chem. A. 1998;102:6957–6966. doi: 10.1021/jp981809p. [DOI] [Google Scholar]
- 27.Pépin C, Goulet T, Houde D, Jay-Gerin JP. Femtosecond kinetic measurements of excess electrons in methanol: substantiation for a hybrid solvation mechanism. J. Phys. Chem. 1994;98:7009–7013. doi: 10.1021/j100079a020. [DOI] [Google Scholar]
- 28.Silva C, Walhout PK, Reid PJ, Barbara PF. Detailed investigations of the pump-probe spectroscopy of the equilibrated solvated electron in alcohols. J. Phys. Chem. A. 1998;102:5701–5707. doi: 10.1021/jp980861i. [DOI] [Google Scholar]
- 29.Soroushian B, et al. Solvation dynamics of the electron produced by two-photon ionization of liquid polyols. 1. Ethyl glycol. J. Phys. Chem. A. 2006;110:1705–1717. doi: 10.1021/jp0540149. [DOI] [PubMed] [Google Scholar]
- 30.Bonin J, Lampre I, Pernot P, Mostafavi M. Solvation dynamics of electron produced by two-photon ionization of liquid polyols. II. Propanediols. J. Phys. Chem. A. 2007;111:4902–4913. doi: 10.1021/jp068323q. [DOI] [PubMed] [Google Scholar]
- 31.Turi L, Rossky PJ. Theoretical studies of spectroscopy and dynamics of hydrated electrons. Chem. Rev. 2012;112:5641–5674. doi: 10.1021/cr300144z. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz BJ, Rossky PJ. Aqueous solvation dynamics with a quantum mechanical solute: computer simulation studies of the photoexcited hydrated electron. J. Chem. Phys. 1994;101:6902. doi: 10.1063/1.468319. [DOI] [Google Scholar]
- 33.Zharikov AA, Fischer SF. Theory of electron solvation in polar liquids: a continuum model. J. Chem. Phys. 2006;124:054506. doi: 10.1063/1.2165198. [DOI] [PubMed] [Google Scholar]
- 34.Elkins MH, Williams HL, Shreve AT, Neumark DM. Relaxation mechanism of the hydrated electron. Science. 2013;342:1496–1499. doi: 10.1126/science.1246291. [DOI] [PubMed] [Google Scholar]
- 35.Karashima S, Yamamoto Y, Suzuki T. Resolving non-adiabatic dynamics of hydrated electrons using ultrafast photoemission anisotropy. Phys. Rev. Lett. 2016;116:137601. doi: 10.1103/PhysRevLett.116.137601. [DOI] [PubMed] [Google Scholar]
- 36.Nordlund D, et al. Probing the electron delocalization in liquid water and ice at attosecond time scales. Phys. Rev. Lett. 2007;99:217406. doi: 10.1103/PhysRevLett.99.217406. [DOI] [PubMed] [Google Scholar]
- 37.Stahler J, Deinert JC, Wegkamp D, Hagen S, Wolf M. Real-time measurement of the vertical binding energy during the birth of a solvated electron. J. Am. Chem. Soc. 2015;137:3520–3524. doi: 10.1021/ja511571y. [DOI] [PubMed] [Google Scholar]
- 38.Savolainen J, Uhlig F, Ahmed S, Hamm P, Jungwirth P. Direct observation of the collapse of the delocalized excess electron in water. Nat. Chem. 2014;6:697–701. doi: 10.1038/nchem.1995. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y, et al. Direct measurement of vertical binding energy of a hydrated electron. Phys. Chem. Chem. Phys. 2010;12:3653–3655. doi: 10.1039/b925741a. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Adhikary A, Shamoun L, Sevilla MD. Do solvated electrons (eaq–) reduce DNA bases? A Gaussian 4 and density functional theory-Molecular dynamics study. J. Phys. Chem. B. 2016;120:2115–2123. doi: 10.1021/acs.jpcb.5b11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth M, Kohanoff J. Excess electron localization in solvated DNA bases. Phys. Rev. Lett. 2011;106:238108. doi: 10.1103/PhysRevLett.106.238108. [DOI] [PubMed] [Google Scholar]
- 42.Pernot, P. ppernot/SK-Ana: Corrects library error in beta version. Zenodo, 10.5281/zenodo.1064370 (2017).
- 43.Takeda N, Poliakov PV, Cook AR, Miller JR. Faster dissociation: measured rates and computed effects on barriers in aryl halide radical anions. J. Am. Chem. Soc. 2004;126:4301–4309. doi: 10.1021/ja0389671. [DOI] [PubMed] [Google Scholar]
- 44.Kočišek J, Pysanenko A, Fárník M, Fedor J. Microhydration prevents fragmentation of uracil and thymine by low-energy electrons. J. Phys. Chem. Lett. 2016;7:3401–3405. doi: 10.1021/acs.jpclett.6b01601. [DOI] [PubMed] [Google Scholar]
- 45.Gu J, Xie Y, Schaefer HF. Glycosidic bond cleavage of pyrimidine nucleosides by low-energy electrons: a theoretical rationale. J. Am. Chem. Soc. 2005;127:1053–1057. doi: 10.1021/ja0400990. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Sevilla MD. The role of πσ* excited states in electron-Induced DNA strand break formation: a time-dependent density functional theory study. J. Am. Chem. Soc. 2008;130:2130–2131. doi: 10.1021/ja077331x. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Sevilla MD. Role of excited states in low-energy electron (LEE) induced strand breaks in DNA model systems: influence of aqueous environment. Chem. Phys. Chem. 2009;10:1426–1430. doi: 10.1002/cphc.200900025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar, A. & Sevilla, M. D. in Handbook of Computational Chemistry (eds. Leszczynski, J. et al.) 1–63 (Springer Science+Business Media, Dordrecht, 2015).
- 49.Schulz GJ. Resonances in electron impact on diatomic molecules. Rev. Mod. Phys. 1973;45:423–486. doi: 10.1103/RevModPhys.45.423. [DOI] [Google Scholar]
- 50.Frisch, M. J. et al. Gaussian v.16 (Gaussian Inc., Wallingford, CT, 2009).
- 51.Li X, Cai Z, Sevilla MD. DFT calculations of the electron affinities of nucleic acid bases: dealing with negative electron affinities. J. Phys. Chem. A. 2002;106:1596–1603. doi: 10.1021/jp013337b. [DOI] [Google Scholar]
- 52.Wang W, Sevilla MD. Protonation of nucleobase anions in gamma-Irradiated DNA and model systems. Which DNA base is the ultimate sink for the electron? Radiat. Res. 1994;138:9–17. doi: 10.2307/3578841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.