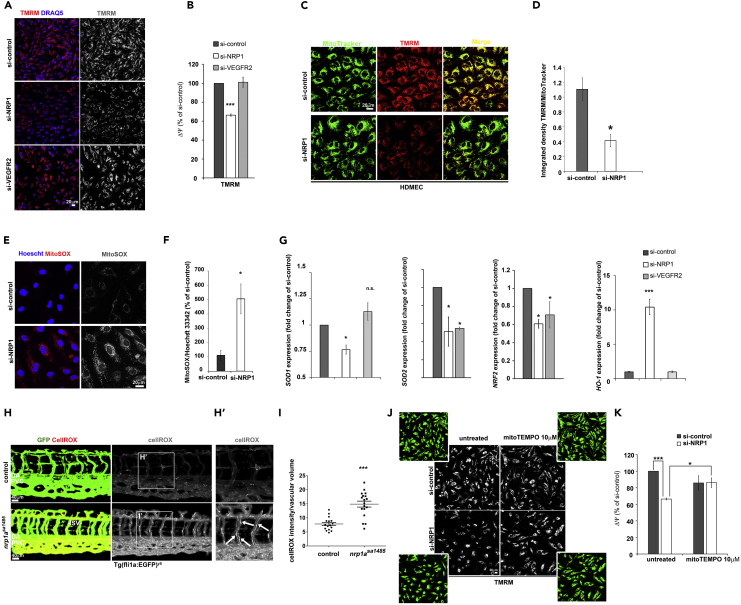
Figure 2.
NRP1 Regulates Mitochondrial Activity and Suppresses Oxidative Stress
(A) HDMECs transfected with si-NRP1, si-VEGFR2, or si-control were incubated with 100 nM TMRM and DRAQ5 (blue) and live imaged with a confocal microscope; scale bar, 20 μm.
(B) Integrated density of TMRM signal per cell was quantified and expressed as percentage (mean ± SEM; n = 4) of si-control.
(C) HDEMCs transfected with siNRP1 or si-control were incubated with 100 nM TMRM and 300 nM MitoTracker Deep Red FM; scale bar, 20 μm.
(D) TMRM and MitoTracker Deep Red FM integrated density was calculated, and the TMRM/MitoTracker ratio visualized in the graph (mean ± SEM; n = 4).
(E) HDMECs transfected with si-NRP1 or si-control were incubated with 5 μM MitoSOX (red and gray) and counterstained with Hoescht 33342 (blue); scale bar, 20 μm.
(F) Integrated density of the MitoSOX signal per cell was quantified and expressed as percentage relative to si-control (mean ± SEM; n = 3).
(G) Quantification of superoxide dismutase-1 and 2 (SOD1, SOD2), NRF2, and heme oxygenase-1 (HO-1) mRNA by RT-qPCR in HDMECs si-NRP1 or si-control expressed as fold change of si-control (mean ± SEM; n = 3).
(H) Tg(fli1a:egfp)y5 wild-type and nrp1asa1485 zebrafish embryos at 3 dpf were incubated with 2.5 μM CellROX (gray) in 10% DMSO for 45 min before imaging; scale bars, 40 μm. (H′) Higher-magnification images of area indicated in the square box in H; white arrows indicate ECs in ISV with high CellROX staining.
(I) CellROX mean intensity was quantified in optical z stacks in n ≥ 16 embryos from three independent experiments after applying a mask to quantify CellROX-positive areas and volumes (mean ± SEM) within the vasculature.
(J) HDMECs treated with mitoTEMPO 10 μM for 24 hr after 48 hr from transfection with si-NRP1 or si-control were incubated with 100 nM TMRM (gray); scale bar, 20 μm.
(K) Integrated density of the TMRM signal per cell was quantified and normalized to cell number determined by manual counting using high-contrast images (panel J, green images) and expressed as percentage relative to si-control (mean ± SEM; n = 3).
*p < 0.05, ***p < 0.001; Student's t test. See also Figure S2).