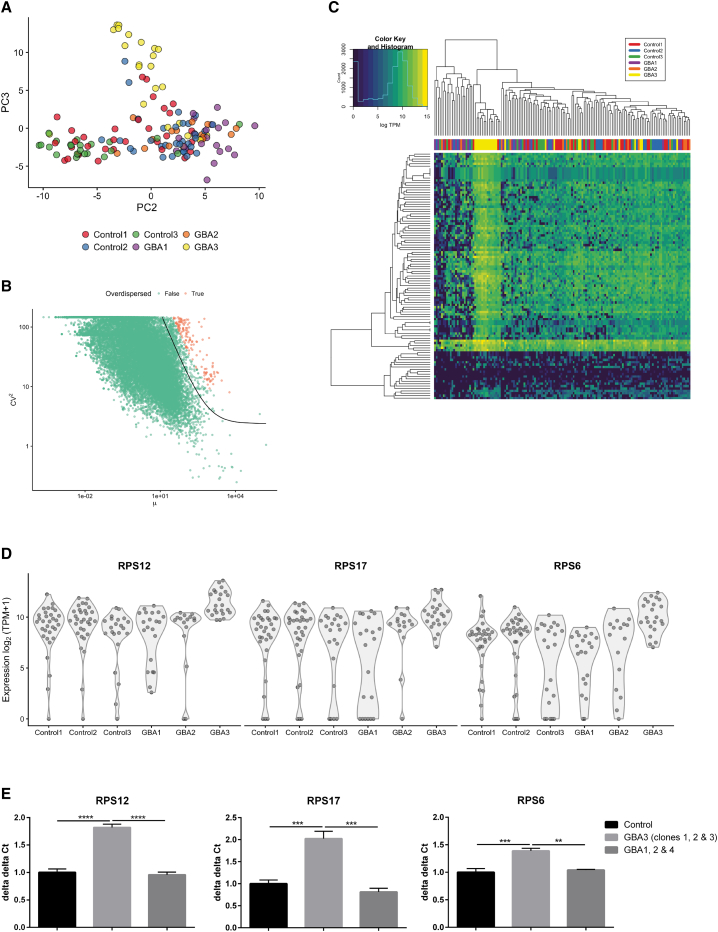
Figure 2.
Single-Cell RNA-Seq Stratification Identifies iPSC-Derived Dopamine Neurons from GBA3 as Significantly Different from Both PD Patient and Control Neurons
(A) Transcriptome PCA analysis resolves GBA3 neurons (yellow) from the remaining two PD GBA-N370S patients and three controls.
(B) Over-dispersion analysis identifies a subset of genes that vary more than expected due to technical fluctuations in the dataset alone.
(C) Heatmap of the single-cell RNA-seq samples identifies an enrichment in the endoplasmic reticulum (ER) signal recognition particle (SRP) pathway in GBA3.
(D) Expression in log2 (TPM+1) of three genes (RPS12, RPS17, and RPS6) prioritized from those significantly DE within the SRP pathway between GBA3 and controls 1, 2, and 3 and GBA1 and 2. DE analysis was performed using a two-sided Wilcoxon signed-rank test on all genes in the SRP pathway.
(E) The upregulation of the three selected genes involved in this pathway was confirmed in iPSC-derived dopamine neurons differentiated from three iPSC clones of GBA3 compared to the three original controls and two PD GBA-N370S patients (GBA1 and 2), plus a fourth PD GBA-N370S patient (GBA4). Data are represented as mean ± SD (∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).