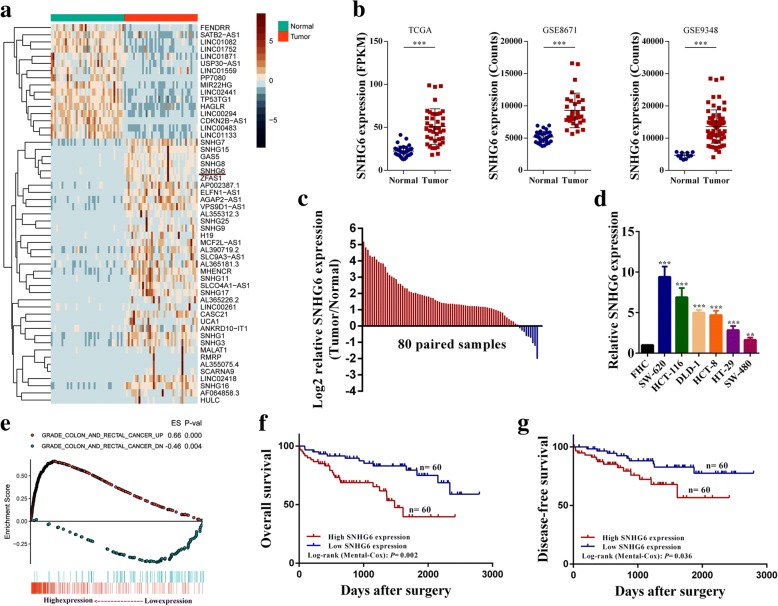
Fig. 1.
SNHG6 expression is upregulated in colorectal cancer and high SNHG6 expression predicts poor prognosis. a Hierarchical cluster heat map of aberrantly expressed lncRNAs in CRC generated from RNA sequencing data from the TCGA database. Red in the heat map denotes upregulation, while blue denotes downregulation. The red line indicates SNHG6. b Expression of SNHG6 in TCGA, GSE8671, and GSE9348 CRC cohorts. c qRT-PCR analysis of SNHG6 expression in 80 pairs of colorectal cancer and corresponding normal tissues. d SNHG6 expression in CRC cell lines (DLD-1, HCT-116, HT-29, SW-620, HCT-8, and SW-480) compared with normal colorectal epithelial cell line FHC detected by qRT-PCR. e Gene set enrichment analysis results of were plotted to visualize the correlation between the expression of SNHG6 and genes dysregulated in colorectal cancer (GRADE_COLON_AND_RECTAL_CANCER_DN and GRADE_COLON_AND_RECTAL_CANCER_UP). f Kaplan–Meier survival analysis of CRC patients’ overall survival based on their SNHG6 expression, a tissue sample whose threshold cycle (CT) value of SNHG6 minus CT value of GAPDH less than − 5.56 was defined as high SNHG6 expression (n = 120, P = 0.002). g Kaplan–Meier survival analysis of CRC patients’ disease-free survival based on their SNHG6 expression, a tissue sample whose threshold cycle (CT) value of SNHG6 minus CT value of GAPDH less than − 5.56 was defined as high SNHG6 expression (n = 120, P = 0.036). **P < 0.01 and ***P < 0.001