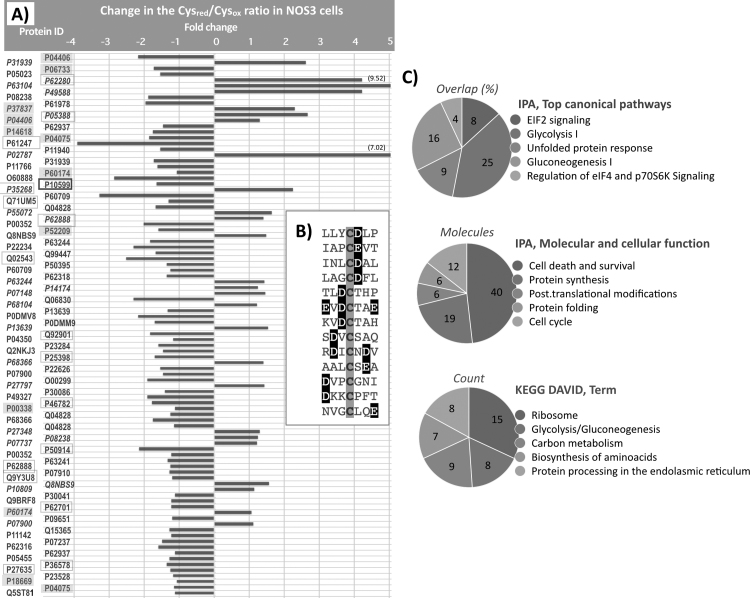
Fig. 2.
NOS3 over expression alters the Reduced/Oxidized ratio of specific Cys on target proteins. A) The UniProt ID of the protein is shown in the vertical axis and the relative change in the Cysred/Cysox ratio in 4TO_NOS cells is indicated in the horizontal axis as fold change relative to the ratio in control 4TO cells transformed with the empty vector. Proteins are ordered according to their statistical q score (Storey & Tibshirani) from 0.0000516 (top) to 0.0497383 (bottom). Proteins with negative values were more oxidized after NOS3 overexpression; those with positive values (IDs in italic) were more reduced. Two proteins showed reductive changes higher than fivefold as indicated. P10599, square boxed with thick line, is Trx1(C73); ID of enzymes of Glycolysis and its branching pathways are highlighted with grey squares and ribosomal proteins are square boxed with thin line. B) sequence context around the cysteine residue of 13 peptides whose reduced/oxidized ratio increased ≥ 1.5 on NOS3 overexpression, showing the presence of acidic residues (white letters highlighted in black) close to the affected cysteine (grey shadowed). Protein names and Cys mapping are detailed in sheet “NOS3 Vo” of the Excel file shown in Suppl Fig. 2. C) Systems Biology analysis of the whole set of redox modified proteins using Ingenuity Pathway Analysis (Qiagen) and DAVID [Huang et al. Nature Protoc. 2009;4(1):44–57]. The five most significant terms from each analysis are shown with p values ranging from 9.18E−19 to 9.75E−04.