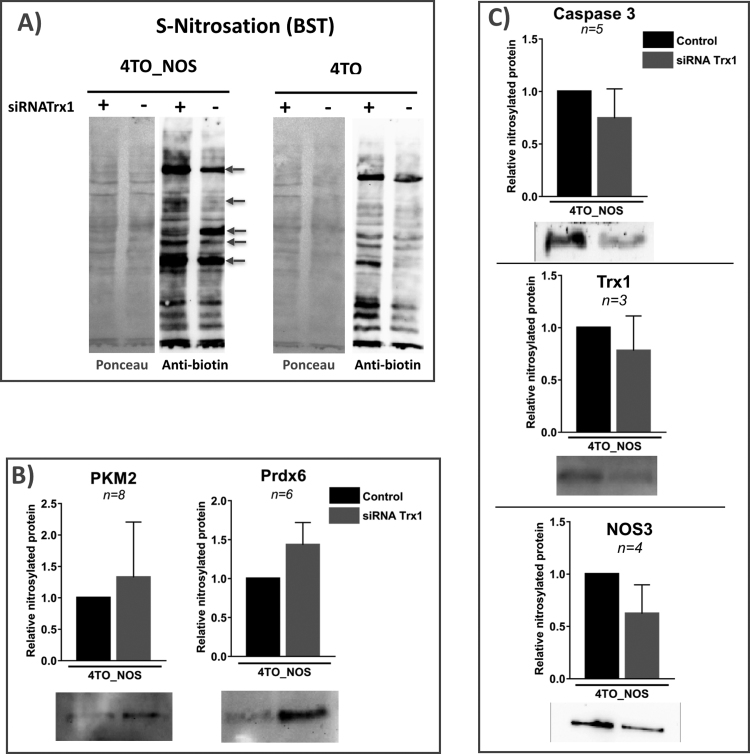
Fig. 4.
Protein S-nitrosation in HepG2 cells over expressing NOS3 and treated with specific siRNA-Trx1. A) NOS3 over expressing HepG2 cells, 4TO-NOS, and control 4TO cells transformed with empty vector were silenced or not, for Trx1 and subject to BST; biotinylated proteins constituting the “S-nitrosome” were separated on SDS-PAGE, transferred to Nitrocellulose membrane and revealed with avidin-peroxidase conjugate. Membranes stained with Ponceau reagent before transfer are presented in the left panels to show the protein load. Arrows point to bands with increased or diminished intensity upon Trx1 silencing. B) and C) biotinylated proteins from 4TO-NOS HepG2 cells were captured on avidin-agarose, eluted with ß-ME, subjected to Western blot and revealed with specific antibodies to 5 selected proteins as indicated. The intensity of the bands was quantified by image analysis as described in M&M; relative abundances of each protein in the fraction of biotinylated proteins compared to non-silenced control cells are shown, together with the number of replicas and the bars showing standard deviation; trimmed images from membranes showing representative band patterns are included below each graph.