Figure 1.
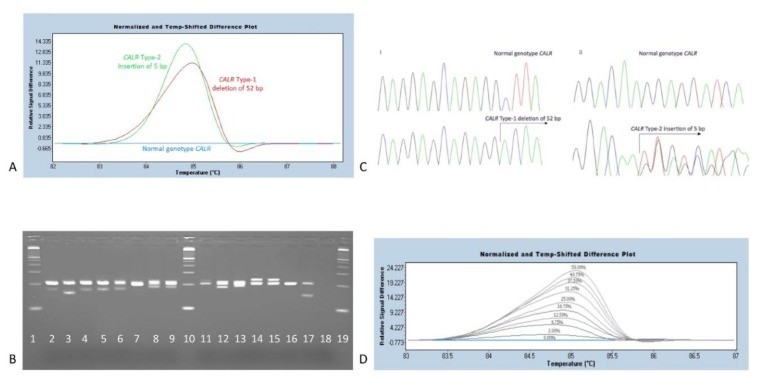
A. HRM-A curves after normalization of temperature and fluorescence data, using the LightCycler 480 SW 1.5.1 software (ROCHE Diagnostics, Indianapolis, IN, US). The two most common CALR exon 9 mutations detected, are Type-1 deletion (L367fs*46) and Type-2 insertion (K385fs*47). These are grouped separately, due to variations in the pattern of their melting curves. Both melting curves differ significantly compared to the normal genotype sample. Each Type-1-like, Type-2-like and complex mutation identified provided a unique melting curve pattern, that diverges from a normal control. B. Agarose gel 3% dense electrophoresis of PCR products. Left to right: line 1, MW marker; line 2, a 34 bp deletion (E364fs*55); line 3, a Type-1 deletion of 52 bp (L367fs*46); line 4, a 37 bp deletion (L367fs*50); line 5, a 34 bp deletion (L367fs*52); line 6, a 31 bp deletion (D373fs*47); line 7, a complex mutation (D373fs*56); line 8, a complex mutation (D373fs*51); line 9, a 19 bp deletion (K375fs*49); line 10, the MW marker; line 11, a complex mutation (K377fs*55); line 12, a 19 bp deletion (K377fs*47); line 13, a complex mutation (E379fs*47); line 14, a 5 bp insertion (E386fs*46); line 15, a Type-2 insertion of 5 bp (K385fs*47); line 16, a normal CALR genotype sample; line 17, a MARIMO cell line DNA sample carrying a 61 bp deletion (L367fs*43) which was initially used as a study reference; line 18, a Non Template Control sample (NTC); line 19 the MW marker. A 100 bp molecular-weight size marker (MW) was used as a DNA ladder (Thermo-Fischer Scientific). C. Sanger sequencing chromatograms for sequence identification of the HRM-A previously detected CALR mutations. i. Top to bottom: a CALR normal genotype sample and a common Type-1 deletion (L367fs*46). ii. Top to bottom: a CALR normal genotype sample and a Type-2 insertion (K385fs*47). D. LoD analysis of the designed HRM-A protocol. A sample harboring a Type-1 deletion mutation (L367fs*46) was used as a reference. Starting at 50% of mutation allele burden, as determined after Sanger sequencing analysis, we ended up detecting as low as 2% of mutant alleles, through serial dilutions.