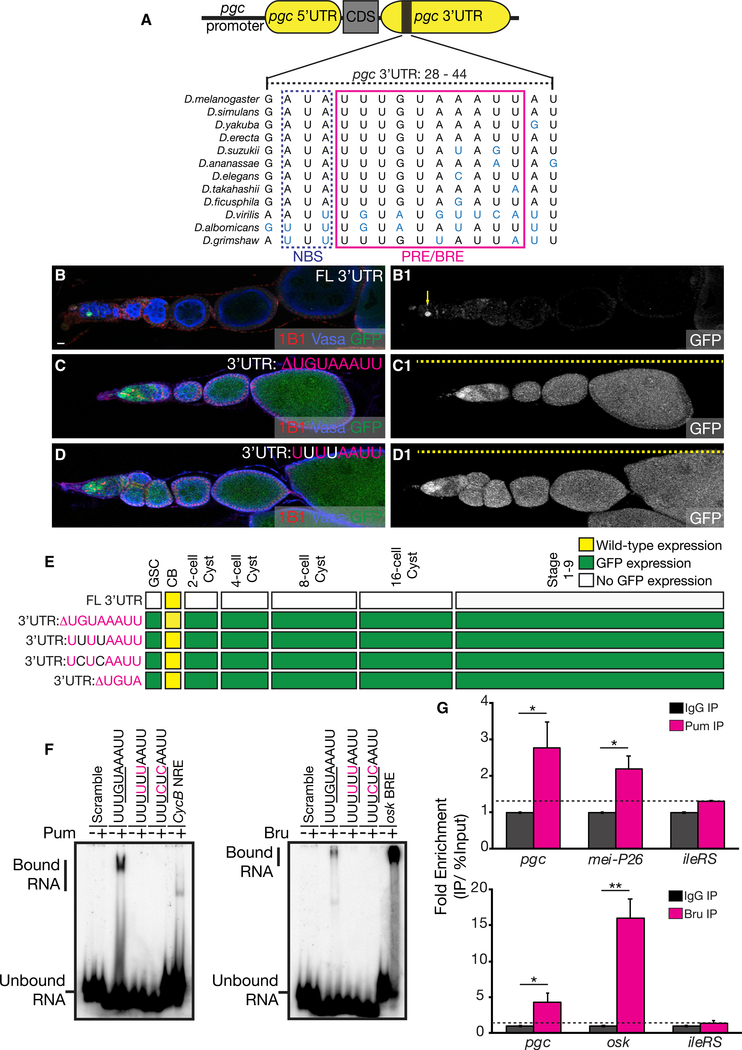
Figure 2. A cis-Element in the pgc 3′ UTR that Binds Pum and Bru Is Required for Translational Control throughout Oogenesis.
(A) The NBS and PRE and/or BRE sequence identified in the pgc 3′ UTR is conserved in 12 species of Drosophilids.
(B) An ovariole of a pgcGFP fly stained with 1B1 (red), Vasa (blue), and GFP (green) showing that GFP expression is restricted to the pre-CB (arrow in B1).
(C) An ovariole of a pgcGFP reporter that lacks the PRE and/or BRE sequence in the 3′ UTR stained with 1B1 (red), Vasa (blue), and GFP (green). GFP regulation was lost throughout oogenesis (dashed line in C1).
(D) An ovariole of a pgcGFP reporter in which the PRE and/or BRE core UGUA motif was mutated stained with 1B1 (red), Vasa (blue), and GFP (green). GFP regulation was lost throughout oogenesis (dashed line in D1).
(E) A developmental profile of GFP expression in different stages of oogenesis of transgenes in which the PRE and/or BRE sequence was either deleted ormutated.
(F) EMSAs show that purified Pum and Bru proteins bind to the PRE and/or BRE of the pgc 3′ UTR, the NRE of the CycB 3′ UTR, and the BRE of the osk 3′ UTR, respectively.
(G) qPCR of pgc, mei-P26 (positive control), and ileRS (negative control) carried out on RNA samples extracted after an IP with Pum antibody (top). qPCR of pgc, osk (positive control), and ileRS (negative control) carried out on RNA samples extracted after an IP with Bru antibody (bottom). RIP-qPCR graphs represent an average generated from three independent biological samples. The error bars represent SE. A Student’s t test analysis was performed. * and ** indicate a p value < 0.05 and < 0.005, respectively.
Scale bar, 10 μm. See also Figures S1 and S2.