Extended Data Fig.7. Transcriptional regulation of g-beige adipocyte differentiation.
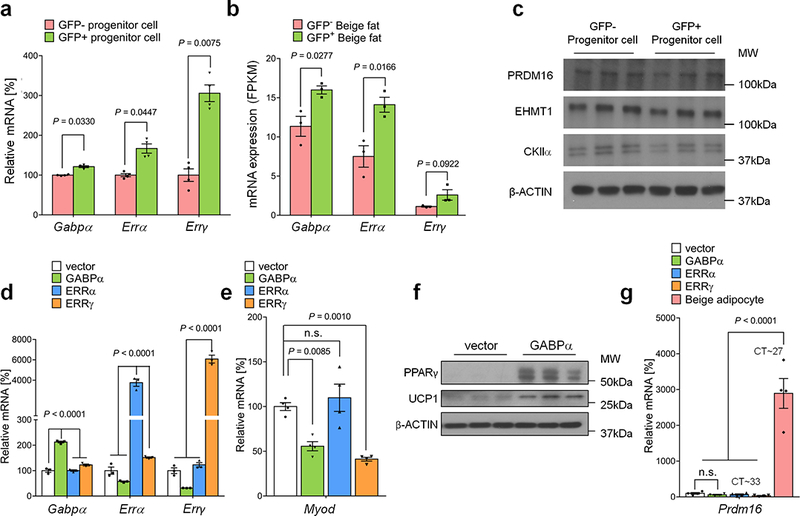
a, mRNA expression of Gabpα, Errα, and Errγ in GFP+ and GFP- progenitors in the inguinal WAT of Myod-CreERT2 reporter mice treated with β-blocker. n=4. b, mRNA expression of indicated genes in GFP+ and GFP- beige fat from Myod-CreERT2 reporter mice. n=3. c, Protein expression of PRDM16, EHMT1, and CKIIα in GFP+ and GFP- progenitors in the inguinal WAT of Myod-CreERT2 reporter mice. β-actin was used as a loading control. Molecular weight (kDa) is shown on the right. d, mRNA expression of indicated genes in C2C12 myoblasts expressing an empty vector, GABPα, ERRα, or ERRγ by lentivirus. n=3. e, mRNA expression of Myod in C2C12 myoblasts expressing an empty vector, GABPα, ERRα, or ERRγ by lentivirus. n.s., not significant. n=4. f, Protein expression of PPARγ and UCP1 in differentiated C2C12 cells expressing an empty vector or GABPα under pro-adipogenic conditions. β-actin was used as a loading control. Molecular weight (kDa) is shown on the right. The blots represent five independent samples. g, mRNA expression of Prdm16 in C2C12 myoblasts expressing an empty vector, GABPα, ERRα, or ERRγ by lentivirus. Differentiated immortalized beige adipocytes are included as a reference. n=4. (a,b) Data are mean ± SEM of biologically independent replicates, and analyzed by unpaired two-sided Student’s t-test. (d,e,g) Data are mean ± SEM of biologically independent replicates, and analyzed by ANOVA followed by Tukey’s test.
