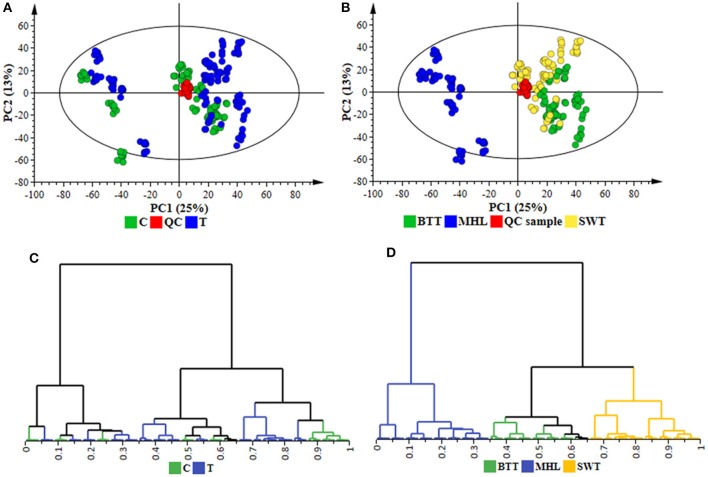
Figure 2.
Unsupervised chemometric modeling (ESI negative data): (A) A PCA scores scatter plot of all the samples, including the QC samples, colored according to the treatment (C = control, T = treated). (B) The scores plot in (A) but colored according to cultivars (MHL = Mhlophe, BTT = NS 5511, SWT = NS 5655). The PCA model presented here was an 18-component model (of the Pareto-scaled data matrix X), with R2 of 0.739, explained variation, and Q2 of 0.664, predicted variation, according to seven-fold cross validation. (C,D) are HCA dendrograms corresponding to (A,B), respectively. The unsupervised modeling provided a global overview of the data shown in the PCA scores plots and HCA dendrograms, allowing the identification of sample grouping and natural clustering with regards to treatment-related and cultivar-dependent groupings (A–D).